1. Introduction: Transcription, the Big Picture
In a previous tutorial, we covered transcription in prokaryotes. Before we look at transcription in eukaryotes, let’s review.
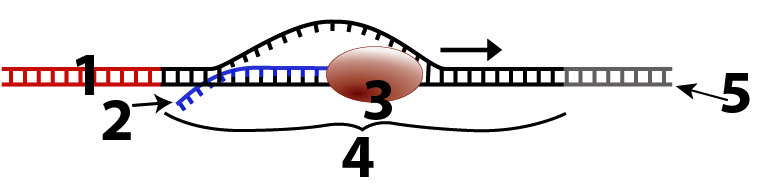
RNA polymerase (3, below) is the enzyme that transcribes DNA into RNA. To do so, it starts by binding at a promoter (1). The promoter is a sequence of DNA that signals to RNA polymerase that this is where transcription should start. Once joined to the promoter, RNA polymerase transcribes the template DNA (at 5) into RNA (at 2). Transcription continues until the RNA polymerase reaches a terminator sequence (which is not shown in the diagram below).
In eukaryotes, this system has several additional features.
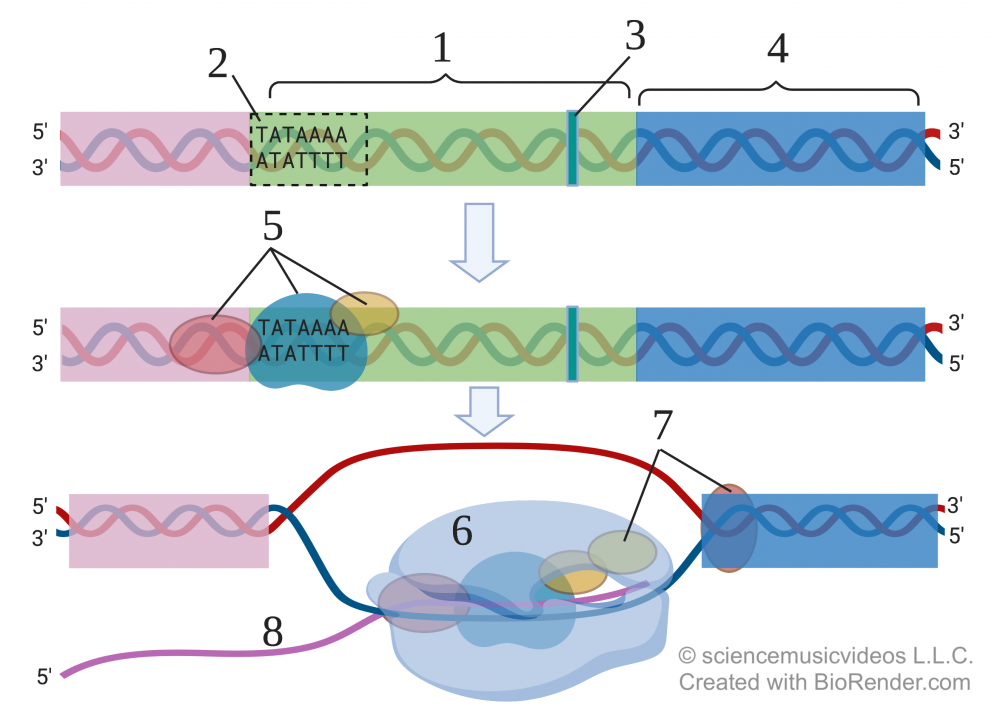
Eukaryotic promoters (1) usually begin with a highly conserved DNA sequence called a TATA box (2). The name comes from the fact that this sequence, found in many eukaryotic genes, begins with TATAAA. Between 25 and 35 bases later is the point where transcription starts (the first DNA base that will be transcribed into RNA). This is indicated by number 3. Following the promoter is the template DNA (at 4). This is the DNA that gets transcribed into RNA.
Before RNA polymerase can bind with the promoter, a team of general transcription factors (5) has to bind with the promoter. General transcription factors bind to most promoters, often at the TATA Box.
Once these general transcription factors are in place, RNA polymerase (6) binds. But transcription won’t begin until additional transcription factors (7) bind. Once all of this is in place, RNA polymerase will begin to transcribe the DNA into RNA (8).
Let’s quickly check to see if you understand the two diagrams above.
2. Checking Understanding: Transcription and the Transcription Initiation Complex
[qwiz random = “true” qrecord_id=”sciencemusicvideosMeister1961-M24_transcription initiation complex”] [h]
Transcription and the transcription initiation complex
[i]
[q] In the diagram below, number 1 is the [hangman]
[c]IHByb21vdGVy[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, number 3 is RNA [hangman]
[c]IHBvbHltZXJhc2U=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, the TATA box is at
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIEEtVC1yaWNoIHJlZ2lvbiBhdCB0aGUgc3RhcnQgb2YgdGhlIHByb21vdGVyIGlzIGtub3duIGFzIHRoZSBUQVRBIGJveC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgVEFUQSBib3ggaXMgcmljaCBpbiBhZGVuaW5lIGFuZCB0aHltaW5lIGJhc2UgcGFpcnMu[Qq]
[q] In the diagram below, the template DNA is at
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IE5pY2Ugam9iLiAmIzgyMjA7NCYjODIyMTsgaXMgdGhlIHRlbXBsYXRlIEROQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgdGVtcGxhdGUgRE5BIGlzIHRoZSBzdHJhbmQgb2YgRE5BIHRoYXQmIzgyMTc7cyB0cmFuc2NyaWJlZCBpbnRvIFJOQS4gSWYgOCBpcyBSTkEsIHRoZW4gd2hhdCBoYXMgdG8gYmUgdGhlIHRlbXBsYXRlIHN0cmFuZCBvZiBETkE/[Qq]
[q] In the diagram below, a general transcription factor is shown at
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IFdheSB0byBnby4gJiM4MjIwOzUmIzgyMjE7IGlzIGEgZ2VuZXJhbCB0cmFuc2NyaXB0aW9uIGZhY3Rvci4=[Qq]
[c]IDc=[Qq]
[f]IE5vLCBidXQgeW91JiM4MjE3O3JlIG9uIHRoZSByaWdodCB0cmFjay4gJiM4MjIwOzcmIzgyMjE7IGlzIGEgdHJhbnNjcmlwdGlvbiBmYWN0b3IsIGJ1dCBpdCYjODIxNztzIGJpbmRpbmcgZmFpcmx5IGxhdGUgaW4gdGhlIHByb2Nlc3MuIEZpbmQgYSB0cmFuc2NyaXB0aW9uIGZhY3RvciB0aGF0IHNob3dzIGJpbmRpbmcgYmVmb3JlIFJOQSBwb2x5bWVyYXNlIGFycml2ZXMu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHNvbWV0aGluZyB0aGF0IGJpbmRzIHRvIHRoZSBETkEgYmVmb3JlIFJOQSBwb2x5bWVyYXNlIChhbmQgbWFrZXMgdGhhdCBiaW5kaW5nIHBvc3NpYmxlKS4=[Qq]
[q] In the diagram below, RNA polymerase is shown at
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]IE5pY2UuICYjODIyMDs2JiM4MjIxOyBpcyBSTkEgcG9seW1lcmFzZS4=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHNvbWV0aGluZyB0aGF0IGJpbmRzIHRvIHRoZSBETkEgYmVmb3JlIFJOQSBwb2x5bWVyYXNlIChhbmQgbWFrZXMgdGhhdCBiaW5kaW5nIHBvc3NpYmxlKS4=[Qq]
[q] In the diagram below, newly transcribed RNA is shown at
[textentry single_char=”true”]
[c]ID g=[Qq]
[f]IEdvb2Qgam9iLiAmIzgyMjA7OCYjODIyMTsgaXMgbmV3bHkgdHJhbnNjcmliZWQgUk5BLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaW5kIHNvbWV0aGluZyBSTkEgcG9seW1lcmFzZSAoNikgaXMgc3ludGhlc2l6aW5nIGZyb20gdGhlIEROQSB0ZW1wbGF0ZS4=[Qq]
[c]IEVudGVyIGxldHRlcg==[Qq]
[/qwiz]
3. Eukaryotic Transcription Factors and Control of Gene Expression
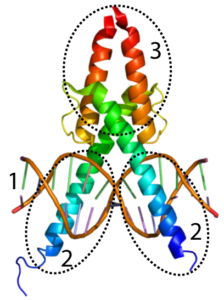
In the section above, you read about the role of transcription factors: molecules that allow RNA polymerase to bind with a promoter and to start transcribing genes. Transcription factors, for the most part, are proteins. A skeletal structure of one transcription factor, MyoD, is shown on the left. MyoD plays a key role in causing certain embryonic cells to develop into muscle tissue.
To do that, MyoD has to bind with DNA. In the diagram to your left, the alpha helices shown at “2” make up a DNA binding domain. Not surprisingly, this DNA binding domain binds with DNA (shown at “1”). The alpha helices at “3” have a different function: they bind with other proteins.
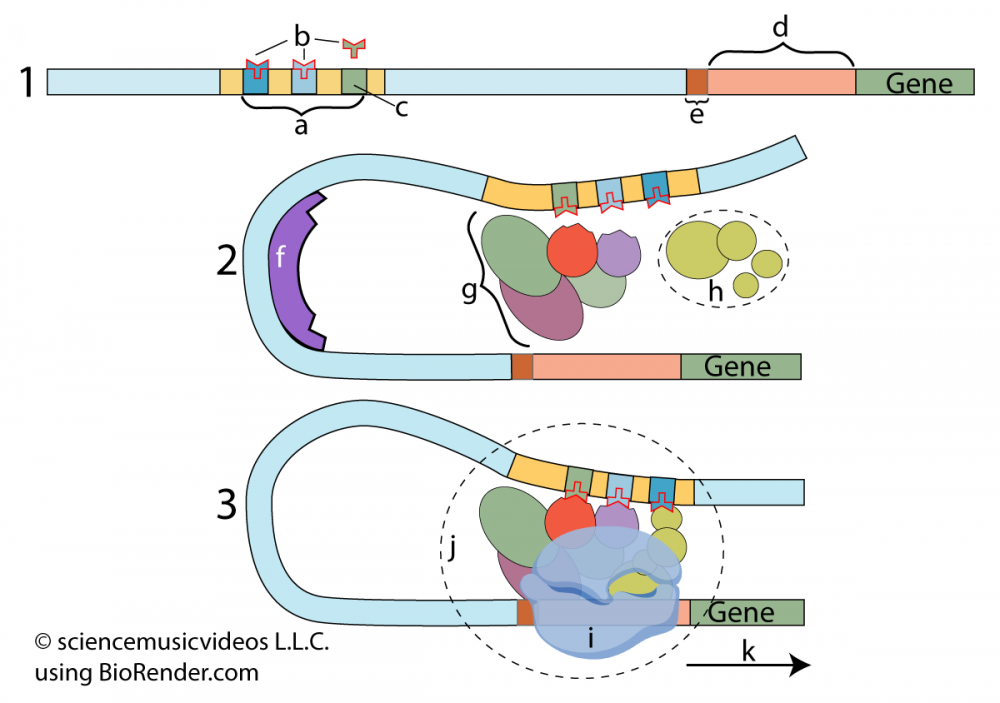 In the series of diagrams to your left, you can see a model that shows these transcription factors at work.
In the series of diagrams to your left, you can see a model that shows these transcription factors at work.
Let’s start with diagram 1: Letter “a” is a region of DNA that acts as a kind of switch that determines whether or not transcription will occur. These regions are called enhancers or regulatory switches. Note that these enhancers can be some distance away from the protein-coding region of the gene.
The enhancer consists of one or more control elements (indicated by “c.”). The transcription factors that bind with these control elements (shown at “b”) are often referred to as activators (because they activate genes).
Now, look at diagram 2. With these activators in place, the enhancer can bind with general transcription factors (at “h”) and mediator proteins (at “g”). If you look carefully at “g” and “h” you can see how their shape was drawn to be complementary to the activators that are bound to the enhancer. On another region of the DNA, a DNA-bending protein (at “f”) bends the DNA in a way that causes the enhancer to approach the promoter.
In diagram 3, all of this comes together to form a transcription initiation complex (j). This allows RNA polymerase (i) to bind with the promoter, enabling transcription (“k”).
What’s the takeaway? For a eukaryotic gene to be transcribed, activator proteins (transcription factors) need to bind with enhancers (regulatory switches). These, in turn, bind with other proteins to form a transcription initiation complex. Once that’s happened, eukaryotic genes can be transcribed into RNA.
[qwiz random = “true” qrecord_id=”sciencemusicvideosMeister1961-M24_Eukaryotic Gene structure and transcriptional contol” style=”width: 650px !important; min-height: 400px !important;”]
[h]Eukaryotic Transcription
[i]
[q] In the diagram below, which number indicates the part of the transcription factor that would interact with another transcription factor?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEV4Y2VsbGVudC4gTnVtYmVyICYjODIyMDszJiM4MjIxOyBpcyBhIHByb3RlaW4gYmluZGluZyBkb21haW4u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBKdXN0IHVzZSBhIHByb2Nlc3Mgb2YgZWxpbWluYXRpb24uIFdoaWNoIHBhcnQgaXNuJiM4MjE3O3Q=IGJpbmRpbmcgd2l0aCBETkE/[Qq]
[q] In the diagram below, which letter indicates an enhancer?
[textentry single_char=”true”]
[c]IG E=[Qq]
[f]IE5pY2Ugam9iISBUaGUgbGV0dGVyICYjODIyMDthJiM4MjIxOyBpbmRpY2F0ZXMgYW4gZW5oYW5jZXIu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgZW5oYW5jZXIgaXMgYSBjb250cm9sIHJlZ2lvbiBsb2NhdGVkIHdheSB1cHN0cmVhbSBmcm9tIHRoZSBnZW5lLCBhbmQgaXQgY29uc2lzdHMgb2YgbXVsdGlwbGUgY29udHJvbCBlbGVtZW50cy4=[Qq]
[q] In the diagram below, which letter indicates an activator?
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IE5pY2Ugam9iLiBMZXR0ZXIgJiM4MjIwO2ImIzgyMjE7IGlzIGFuIGFjdGl2YXRvci4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBBY3RpdmF0b3JzIGFyZSB0cmFuc2NyaXB0aW9uIGZhY3RvcnMgdGhhdCBiaW5kIHdpdGggdGhlIGVuaGFuY2VyIHJlZ2lvbi4=[Qq]
[q] In the diagram below, which letter indicates a control element?
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IEF3ZXNvbWUuIExldHRlciAmIzgyMjA7YyYjODIyMTsgaXMgYSBjb250cm9sIGVsZW1lbnQu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBDb250cm9sIGVsZW1lbnRzIGFyZSBwYXJ0cyBvZiBlbmhhbmNlcnMu[Qq]
[q] In the diagram below, which letter indicates the promoter?
[textentry single_char=”true”]
[c]IG Q=[Qq]
[f]IEdvb2Qgd29yay4gTGV0dGVyICYjODIyMDtkJiM4MjIxOyBpbmRpY2F0ZXMgdGhlIHByb21vdGVyLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgcHJvbW90ZXIgaXMganVzdCB1cHN0cmVhbSBvZiB0aGUgZ2VuZS4=[Qq]
[q] In the diagram below, if “g” indicates mediator proteins. which letter indicates general transcription factors?
[textentry single_char=”true”]
[c]IG g=[Qq]
[f]IFRlcnJpZmljLiBMZXR0ZXIgJiM4MjIwO2gmIzgyMjE7IGluZGljYXRlcyBnZW5lcmFsIHRyYW5zY3JpcHRpb24gZmFjdG9ycy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBGaXJzdCBvZiBhbGwsIHRoZSBxdWVzdGlvbiB0ZWxscyB5b3UgdGhhdCB0aGUgYW5zd2VyIGlzIG5vdCAmIzgyMjA7Zy4mIzgyMjE7IFNlY29uZCwgbG9vayBhdCBkaWFncmFtIG51bWJlciAzLCB3aGVyZSB5b3UgY2FuIHNlZSB0aGUgZ2VuZXJhbCB0cmFuc2NyaXB0aW9uIGZhY3RvcnMgaW50ZXJhY3Rpbmcgd2l0aCBib3RoIHRoZSBwcm9tb3RlciBhbmQgUk5BIHBvbHltZXJhc2UuIElmIGl0JiM4MjE3O3Mgbm90ICYjODIyMDtnLCYjODIyMTsgdGhlbiB3aGF0IG11c3QgaXQgYmU/[Qq]
[q] In the diagram below, if “h” indicates general transcription factors, then what letter has to be mediator proteins?
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IFRlcnJpZmljLiBMZXR0ZXIgJiM4MjIwO2cmIzgyMjE7IGluZGljYXRlcyBtZWRpYXRvciBwcm90ZWlucy4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBNZWRpYXRvciBwcm90ZWlucyBpbnRlcmFjdCB3aXRoIGFjdGl2YXRvcnMgYW5kIHdpdGggZ2VuZXJhbCB0cmFuc2NyaXB0aW9uIGZhY3RvcnMuIFdoYXQgcGFydCBhYm92ZSBpcyBkb2luZyBib3RoPw==[Qq]
[q] In the diagram below, DNA bending proteins are shown at
[textentry single_char=”true”]
[c]IG Y=[Qq]
[f]IE5pY2UuIExldHRlciAmIzgyMjA7ZiYjODIyMTsgaW5kaWNhdGVzIGEgRE5BLWJlbmRpbmcgcHJvdGVpbi4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGNhcmVmdWxseSBhdCB0aGUgZGlhZ3JhbS4gV2hhdCBwYXJ0IHNlZW1zIHRvIGJlIGFzc29jaWF0ZWQgd2l0aCBiZW5kaW5nIHRoZSBETkEgc28gdGhhdCB0aGUgZW5oYW5jZXIgY29tZXMgY2xvc2UgdG8gdGhlIHByb21vdGVyPw==[Qq]
[q] In the diagram below, RNA polymerase is shown at
[textentry single_char=”true”]
[c]IG k=[Qq]
[f]IEdvb2Qgd29yay4gVGhlIGxldHRlciAmIzgyMjA7aSYjODIyMTsgaW5kaWNhdGVzIFJOQSBwb2x5bWVyYXNlLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBzb21ldGhpbmcgdGhhdCBzaXRzIHNxdWFyZWx5IG9uIHRoZSBwcm9tb3RlciAoYW5kIGl0IGxvb2tzIGEgYml0IGxpa2UgYSBndWl0YXIgcGljayku[Qq]
[q] In the diagram below, a transcription initiation complex is shown at
[textentry single_char=”true”]
[c]IG o=[Qq]
[f]IEF3ZXNvbWUuIExldHRlciAmIzgyMjA7aiYjODIyMTsgaW5kaWNhdGVzIHRoZSB0cmFuc2NyaXB0aW9uIGluaXRpYXRpb24gY29tcGxleC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgdHJhbnNjcmlwdGlvbiBpbml0aWF0aW9uIGNvbXBsZXggY29uc2lzdHMgb2YgYWN0aXZhdG9ycywgbWVkaWF0b3JzLCBnZW5lcmFsIHRyYW5zY3JpcHRpb24gZmFjdG9ycywgYW5kIFJOQSBwb2x5bWVyYXNlLCByZWFkeSBmb3IgdHJhbnNjcmlwdGlvbi4=[Qq]
[x]
[restart]
[/qwiz]
4. Once DNA is transcribed, several steps are required before the RNA can be translated into protein
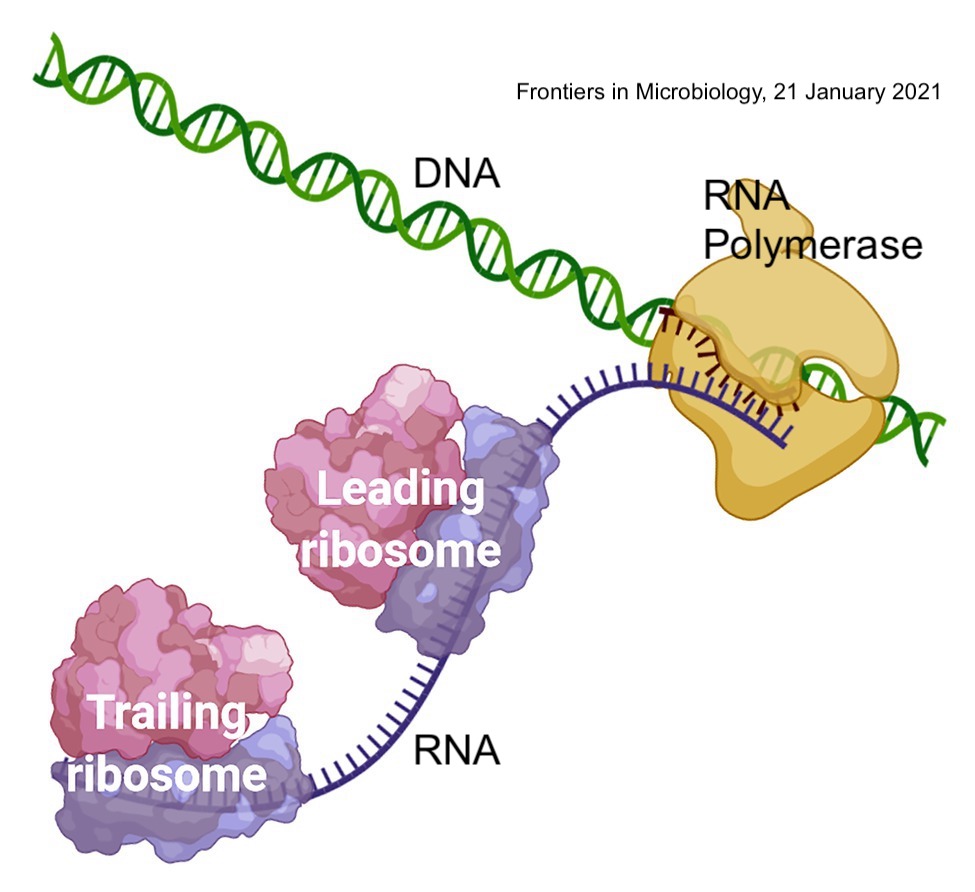
In prokaryotic cells, translation can immediately follow the transcription of RNA from DNA. This is called coupled transcription-translation.
In eukaryotes, several steps are required between the creation of the initial RNA transcript (“2” below) and mRNA that’s ready for translation (“3”).
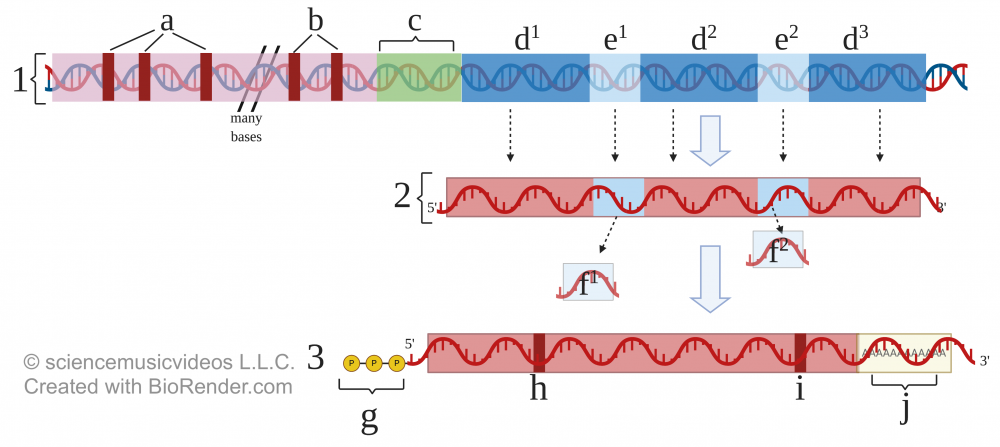
To start, let’s look at the DNA (at 1). The letters “a” and “b” indicate control elements (sequences that activators can bind to). Letter “c” is the promoter. The gene starts at d1 and ends at d3. So what’s going on with e1 and e2?
Protein-coding regions in eukaryotic DNA are interrupted by intervening sequences of non-coding called introns. Imagine that in the middle of a sentence, every few words included a bunch of nonsense letters. Applied to the preceding sentence, here’s what that would look like:
Imagine that in the slkajfpewra;lskdjfweroj middle of a sentence, aklmre1oqw every few words included a bunch peiurpq0erI of nonsense letters.
Eukaryotic genes are similarly interrupted by sequences of nucleotide bases that, if translated, would ruin the function of the protein that the gene is coding for. However, because of a phenomenon called “alternative splicing” (discussed later in this module), these introns are not edited out of the DNA. Consequently, before translating their RNA into protein, eukaryotic cells have to edit their RNA. That editing involves removing introns and leaving in the RNA that will be expressed. Those expressed sequences have a name: exons. Just remember: exons get expressed; introns are intervening segments.
To follow what happens, look again at the “RNA Processing” diagram above. Number 1 shows the template DNA. Letters d1, d2, and d3 represent exon DNA that will become messenger RNA and be coded into protein. But disrupting the exons are two introns, represented by e1 and e2.
Number 2 represents what’s called “pre-mRNA.” This transcribed RNA includes the introns, indicated by f1 and f2.
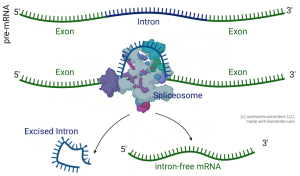
Removing the intron RNA and connecting the exons is called splicing. If you’ve ever worked with a digital video file (or with film or audiotape), you know what splicing is: you cut out the material you don’t want and connect the parts that you do want. In a eukaryotic cell, Small Nuclear RiboNuclear Proteins (abbreviated as “snRNPs” and pronounced as snurps) combine to form a splicing machine called a spliceosome. Spliceosomes can find the introns, cut them out, and connect the exons.
After that, two more steps are required. These are shown in step 3. First, enzymes add a GTP (guanine triphosphate) “cap” to the RNA’s 5′ end (indicated by the letter “g”). At the 3′ end, other enzymes add a “tail” consisting of repeated adenine ribonucleotides. The tail is called a “poly-A tail.” Both the cap and tail help the mRNA resist enzymatic degradation while in the cytoplasm. Finally, “h” and “i” indicate the start and stop codons, respectively.
Got it? Take the quiz below about RNA processing.
5. Quiz: RNA Processing
[qwiz random = “true” style=”width: 650px !important; min-height: 400px !important;” qrecord_id=”sciencemusicvideosMeister1961-RNA Processing (introns, exons)”]
[h]RNA Processing.
[i]
[q] In the diagram below, letters “a” and “b” both represent [hangman] elements, which are binding sites within an [hangman].
[c]Y29udHJvbA==
[c]ZW5oYW5jZXI=[Qq]
[q] In the diagram below, which letter indicates intron DNA (don’t worry about superscripts)?
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IE5pY2Ugam9iLiBUaGUgaW50cm9uIEROQSAoYXQgJiM4MjIwO2UmIzgyMjE7KSBiZWNvbWVzIFJOQSBpbiB0aGUgcHJlLVJOQSwgYnV0IHRoZW4gY3V0cyBjdXQgb3V0IGFzIG1STkEgaXMgZm9ybWVkLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLsKgVGhlIGludHJvbiBETkEgY29kZXMgZm9yIFJOQSB0aGF0IGdldHMgY3V0IG91dCBvZiB0aGUgcHJlLVJOQSBiZWZvcmUgaXQgZ2V0cyBtYWRlIGludG8gbVJOQS4gJiM4MjIwO0YmIzgyMjE7IHJlcHJlc2VudHMgdHdvIFJOQSBzZWdtZW50cyB0aGF0IGhhdmUgYmVlbiBjdXQgb3V0JiM4MjMwO3NvIGp1c3Qgd29yayB1cCBmcm9tIHRoZXJlIGFuZCB5b3UmIzgyMTc7bGwgaGF2ZSB0aGUgYW5zd2VyLsKg[Qq]
[q] In the diagram below, pre-mRNA (still with introns) is represented by the number
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEF3ZXNvbWUuICYjODIyMDsyJiM4MjIxOyBpcyBwcmUtbVJOQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTcGxpY2VkIG91dCBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBhdCAmIzgyMjA7Zi4mIzgyMjE7IA==TG9vayBmb3IgUk5BIHRoYXQgc3RpbGwgY29udGFpbnMgaW50cm9ucy7CoA==wqA=[Qq]
[q] In the diagram below, messenger RNA is represented by the number
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEdvb2Qgd29yay4gJiM4MjIwOzMmIzgyMjE7IGlzIG1lc3NlbmdlciBSTkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBpbiBudW1iZXLCoCAmIzgyMjA7Mi4mIzgyMjE7IA==TG9vayBmb3IgUk5BIGluIHdoaWNoIHRoZSBpbnRyb25zIGhhdmUgYmVlbiBzcGxpY2VkIG91dC4=[Qq]
[q] In the diagram below, the protective, 5′ GTP cap is at
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0cmIzgyMjE7IGlzIHRoZSA1JiM4MjQyOyBHVFAgY2FwLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBHVFAgaXMgYXR0YWNoZWQgdG8gbVJOQSwgYW5kIGl0IGhhcyAzIHBob3NwaGF0ZSBncm91cHMuwqA=[Qq]
[q]In the diagram below, which number represents eukaryotic transcription and translation?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzImIzgyMjE7IGlzIGV1a2FyeW90aWMgdHJhbnNjcmlwdGlvbiBhbmQgdHJhbnNsYXRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSZW1lbWJlciB0aGF0IGluIGV1a2FyeW90ZXMsIFJOQSBoYXMgdG8gYmUgcHJvY2Vzc2VkIGJlZm9yZSBpdCBjYW4gYmUgdHJhbnNsYXRlZC4gSW4gcHJva2FyeW90ZXMsIHRoYXQmIzgyMTc7cyBub3QgbmVjZXNzYXJ5LsKg[Qq]
[q]In the diagram below, which number represents pre-mRNA?
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO2ImIzgyMjE7IGlzIHByZS1tUk5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBQcmUtbVJOQSBjb250YWlucyBpbnRyb25zIGFuZCBsYWNrcyBhIDUmIzgyNDI7IGNhcCBhbmQgYSBwb2x5LUEgdGFpbC7CoA==[Qq]
[/qwiz]
6. Coordinated Control of Gene Expression
Throughout the life of a eukaryotic organism, there are many occasions where genes need to be turned on and off together. Unlike prokaryotic cells, these genes are not clustered together in operons. They may be on different chromosomes altogether. In a multicellular eukaryote, these genes might need to be activated in different cells. The mechanism for turning these separate genes on at once involves shared regulatory sequences that respond to the same transcription factors.
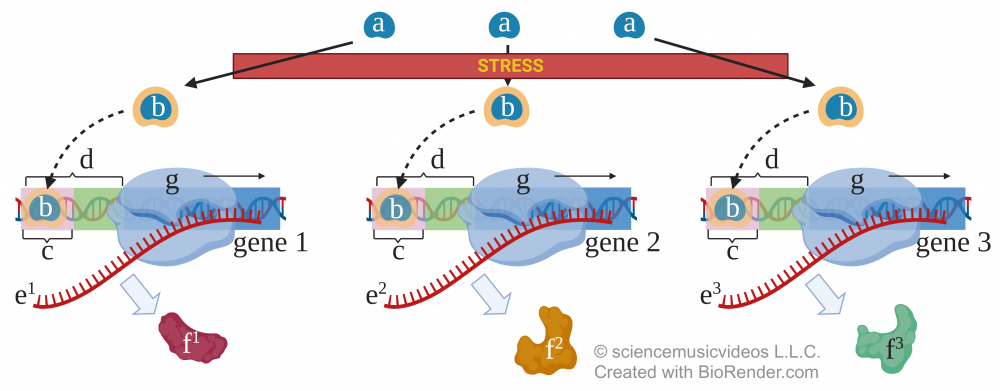
One example of coordinated gene expression involves how plants respond to stress induced by drought. To survive, plants must produce a variety of proteins simultaneously. To do this, inactive transcription factors (shown at “a”) have to respond to stress by changing to an activated form (at “b”). This activation may involve phosphorylation or another type of conformational change.
Once activated, these transcription factors will bind with any promoter that has, as part of its structure, a matching regulatory sequence. In the diagram to the right, you can see three genes, each of which has a promoter (d) containing what’s called a “dehydration response element” (indicated by “c”). The binding of the transcription factor with this element allows RNA polymerase (g) to bind, producing each gene’s RNA (at “e”). This mRNA then gets translated into a variety of proteins (at “f”).
In our bodies (and the bodies of related animals), a similar mechanism explains how one hormone can activate a variety of genes in a variety of cells. An example is the steroid hormone estrogen. Estrogen is known as the feminizing hormone, and in females, it’s involved in the development of secondary sexual characteristics, ovulation, changes in the uterine lining during the menstrual cycle, and sexual receptivity.
Estrogen is produced primarily by ovaries (but also by the placenta during pregnancy). From the ovaries, it diffuses into the bloodstream and then goes everywhere in the body.

As a steroid hormone, estrogen can diffuse through the cell’s phospholipid bilayer. In the cytoplasm, estrogen binds with an estrogen receptor (see the image on right: two molecules of estrogen are circled with a dotted red line).
The estrogen receptor, once bound to estrogen, becomes a transcription factor. It diffuses into the cell’s nucleus, where it binds with estrogen response elements. These control elements combine with other transcription factors to create a transcription initiation complex, resulting in the transcription of estrogen-related genes.
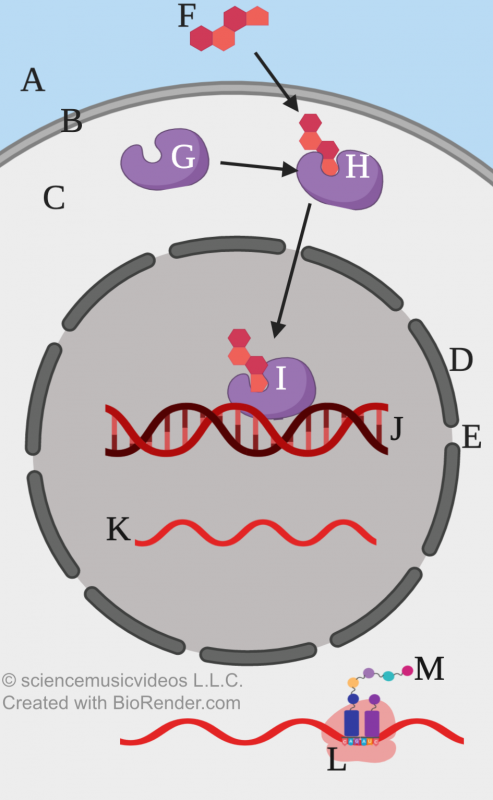
The diagram at left shows a generalized version of this process. “F” represents estrogen. “G” is the estrogen receptor. Once the receptor binds to estrogen, it becomes a transcription factor. Letter “H” shows this transcription factor: a cytoplasmic receptor bound to estrogen. This transcription factor can cross the nuclear membrane (“D”). “I” shows this transcription factor interacting with DNA (“J”), producing RNA that can (after RNA processing) be translated by ribosomes (“L”) into protein (“M”).
7. Cumulative Quiz: Eukaryotic regulation of transcription
[qwiz random = “true” qrecord_id=”sciencemusicvideosMeister1961-M24_activators, cell specialization, coordinated control” style=”width: 650px !important; min-height: 400px !important;”] [h]
Regulation of Eukaryotic Transcription
[q] In the diagram below, the stress is drought. The drought response element is indicated by
[textentry single_char=”true”]
[c]IG M=[Qq]
[f]IE5pY2Ugam9iLiBUaGUgZHJvdWdodCByZXNwb25zZSBlbGVtZW50IGlzIGluZGljYXRlZCBieSAmIzgyMjA7Yy4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBJdCYjODIxNztzIHBhcnQgb2YgdGhlIHByb21vdGVyIHJlZ2lvbiBvZiB0aGUgZ2VuZXMuwqA=[Qq]
[q] In the diagram below, an activated transcription factor is represented by
[textentry single_char=”true”]
[c]IG I=[Qq]
[f]IE5pY2Ugam9iISBMZXR0ZXIgJiM4MjIwO2ImIzgyMjE7IGluZGljYXRlcyBhbiBhY3RpdmF0ZWQgdHJhbnNjcmlwdGlvbiBmYWN0b3Iu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgYWN0aXZhdGVkIHRyYW5zY3JpcHRpb24gZmFjdG9yIGNhbiBiaW5kIHdpdGggKG9yIG5lYXIpIHRoZSBwcm9tb3RlciByZWdpb24u[Qq]
[q] In the diagram below, RNA polymerase is represented by
[textentry single_char=”true”]
[c]IG c=[Qq]
[f]IEV4Y2VsbGVudCEgUk5BIHBvbHltZXJhc2UgaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwO2cuJiM4MjIxOw==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgYWN0aXZhdGVkIHRyYW5zY3JpcHRpb24gZmFjdG9yIGNhbiBiaW5kIHdpdGggKG9yIG5lYXIpIHRoZSBwcm9tb3RlciByZWdpb24u[Qq]
[q] In the diagram below, messenger RNA is represented by
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IEV4Y2VsbGVudCEgTWVzc2VuZ2VyIFJOQSBpcyByZXByZXNlbnRlZCBieSAmIzgyMjA7ZS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMZXR0ZXIgJiM4MjIwO2cmIzgyMjE7IHJlcHJlc2VudHMgUk5BIHBvbHltZXJhc2UmIzgyMTc7cyByb2xlLiBJdCB0cmFuc2NyaWJlcyBETkEgaW50byBSTkEuIElmICYjODIyMDtnJiM4MjIxOyBpcyBSTkEgcG9seW1lcmFzZSwgdGhlbiBtUk5BIG11c3QgYmUgJiM4MjMwOw==[Qq]
[q] In the diagram below, estrogen would be
[textentry single_char=”true”]
[c]IE Y=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtGJiM4MjIxOyByZXByZXNlbnRzIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIGhvcm1vbmUgdGhhdCYjODIxNztzIGNvbWluZyBmcm9tIG91dHNpZGUgdGhlIGNlbGwsIHRoZW4gZGlmZnVzaW5nIGludG8gdGhlIGN5dG9wbGFzbSB0aHJvdWdoIHRoZSBjZWxsIG1lbWJyYW5lLg==[Qq]
[q] In the diagram below, a mobile estrogen receptor before it has bound to estrogen would be represented by
[textentry single_char=”true”]
[c]IE c=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtHJiM4MjIxOyByZXByZXNlbnRzIGEgbW9iaWxlIGVzdHJvZ2VuIHJlY2VwdG9yIGJlZm9yZSBpdCBiaW5kcyB3aXRoIGVzdHJvZ2VuLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIHRoYXQgY2FuIGJpbmQgd2l0aCBlc3Ryb2dlbiAod2hpY2ggaXMgcmVwcmVzZW50ZWQgYnkgJiM4MjIwO0YuJiM4MjIxOyk=[Qq]
[q] In the diagram below, which letter represents the estrogen-receptor complex acting as a transcription factor?
[textentry single_char=”true”]
[c]IG k=[Qq]
[f]IENvcnJlY3QhICYjODIyMDtJJiM4MjIxOyBzaG93cyBhbiBlc3Ryb2dlbi9yZWNlcHRvciBjb21wbGV4IGludGVyYWN0aW5nIHdpdGggRE5BLCBhY3RpbmcgYXMgYSB0cmFuc2NyaXB0aW9uIGZhY3Rvci4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHJlY2VwdG9yIGJvdW5kIHRvIGVzdHJvZ2VuLCBpbnRlcmFjdGluZyB3aXRoIEROQS4=[Qq]
[q] In the diagram below, which letter represents the part that’s translating the transcription product into protein?
[textentry single_char=”true”]
[c]IE w=[Qq]
[f]IENvcnJlY3QhIExldHRlciAmIzgyMjA7TCYjODIyMTsgc2hvd3MgYSByaWJvc29tZS4gdGhlIHJpYm9zb21lIGlzIHRyYW5zbGF0aW5nIFJOQSAodGhlIHRyYW5zY3JpcHRpb24gcHJvZHVjdCkgaW50byBwcm90ZWlu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgdHJhbnNjcmlwdGlvbiBwcm9kdWN0IGlzIFJOQSAoZmlyc3Qgc2hvd24gYXQgJiM4MjIwO0smIzgyMjE7KS4gV2hhdCYjODIxNztzIHRyYW5zbGF0aW5nIHRoZSBSTkEgaW50byBwcm90ZWluPw==[Qq]
[q] In the diagram below, 5 is a general [hangman] factor.
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, number 2 is a [hangman] box.
[c]IFRBVEE=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, number 1 is the [hangman].
[c]IHByb21vdGVy[Qq]
[f]IEdvb2Qh[Qq]
[q] In the diagram below, numbers 5, 6, and 7, arrayed at a gene’s promoter, is called a(n) [hangman] initiation complex.
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, the three DNA elements indicated by “a” together make up an [hangman]. Another word for this same structure is a regulatory [hangman].
[c]IGVuaGFuY2Vy[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IHN3aXRjaA==[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] In the diagram below, the two deleted RNA segments labeled as f1 and f2 must be [hangman]. Once these are removed (and a few other modifications are made), the result (shown at “3”) is [hangman] RNA.
[c]IGludHJvbnM=[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IG1lc3Nlbmdlcg==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, “c” is a control element. Letter “a” is an [hangman]. The molecule that’s binding with “c” is an [hangman].
[c]IGVuaGFuY2Vy[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[c]IGFjdGl2YXRvcg==[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] In the diagram below, “d” is the [hangman]. Letter “f” is a DNA [hangman] protein. Letter “g” is a group of mediator proteins, and “h” is a group of general [hangman] factors. When all of these assemble on the promoter, RNA [hangman] can finally bind and transcribe the gene.
[c]IHByb21vdGVy[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IGJlbmRpbmc=[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IEdvb2Qh[Qq]
[c]IHBvbHltZXJhc2U=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] A key idea represented below is that the action of genes can be [hangman] if these genes share common [hangman] sequences.
[c]IGNvb3JkaW5hdGVk[Qq]
[c]IHJlZ3VsYXRvcnk=[Qq]
[q labels = “top”]
[l]Activators
[fx] No. Please try again.
[f*] Good!
[l]Control Elements
[fx] No. Please try again.
[f*] Good!
[l]DNA Bending protein
[fx] No. Please try again.
[f*] Correct!
[l]Enhancer
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l]Mediator Proteins and Transcription Factors
[fx] No. Please try again.
[f*] Great!
[l]Promoter
[fx] No. Please try again.
[f*] Good!
[l]RNA polymerase
[fx] No, that’s not correct. Please try again.
[f*] Great!
[x][restart]
[/qwiz]