1. Introduction
In the previous tutorials in this module, we’ve looked at how control of eukaryotic genes can involve
- modification of chromatin structure through acetylation and methylation (arrow number 1, above)
- control of transcription (arrow number 2)
In what follows, we’ll look at regulatory methods that follow transcription.
2. Alternative Splicing of RNA
In tutorial 2 of this module, you learned about how eukaryotes need to edit their RNA before it can be translated into protein.
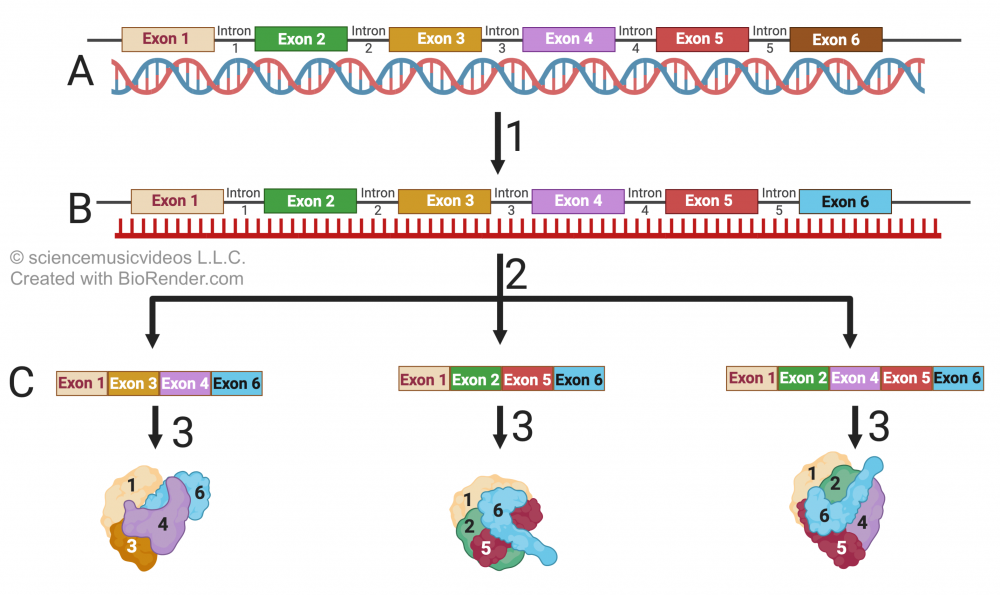 Alternative splicing relates to the fact that exons don’t have to be spliced together in the linear order in which they’re initially transcribed. The order can be changed, and exons can be dropped.
Alternative splicing relates to the fact that exons don’t have to be spliced together in the linear order in which they’re initially transcribed. The order can be changed, and exons can be dropped.
Consider the image at left. In “B” you can see a primary RNA transcript that contains 6 exons. However, alternative splicing (indicated by the arrows at number 2) generates three unique mRNAs, which get translated into three unique proteins, each of which is lacking one or more of the six exons that make up the gene. The result will be three proteins, with different three-dimensional conformations and different properties.
Alternative splicing explains why there are many more human messenger RNAs than there are genes. It’s estimated that about 80% of all human genes are alternatively spliced. Moreover, this might explain some features of human evolution. For example, while the human genome and chimpanzee genome are of equal size, there are significant differences in messenger RNA and protein that are can be attributed to alternative splicing that occurs in humans, and which doesn’t occur in chimpanzees (Principles of Life, Sinauer Associates, p. 222).
3. Regulation of translation by microRNAs (RNA interference)
Protein coding DNA accounts for only about 1.5% of the human genome. What’s the rest of the genome doing? A recent survey of the genome (the ENCODE project) revealed that “more than 80 percent of this non-gene component of the genome, which was once considered “junk DNA,” actually has a role in regulating the activity of particular genes (gene expression)” (see the Genetics home reference). Much of this regulation is thought to happen through non-coding RNAs.
The main type of non-coding RNA to know about is called a microRNA (miRNA). These miRNAs are about 22 nucleotides long. They originate from longer precursor RNAs that are cut apart by an enzyme. After being cut, each 22 nucleotide fragment forms a complex with a protein.
One of the main functions of miRNA is RNA interference. Using complementary base pairing, the miRNA can bind with any cellular RNA (usually messenger RNA) with at least 7 complementary bases. Binding blocks mRNA translation into proteins and is sometimes followed by enzymatic degradation of the mRNA.
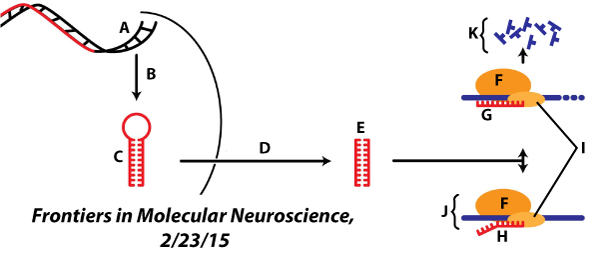
The image at left illustrates microRNA activity. Letter “A” shows non-coding DNA. This is transcribed (“B”) into a pre-microRNA (“C”). Processing and export from the nucleus (“D”) convert this into a microRNA (“E”), which then associates with a protein called an RNA-induced silencing complex (“I”). If the miRNA partially matches its target messenger RNA (as shown at “H”) then translation is blocked (“J”). A complete match (“G”) results in RNA degradation (“K”). Both cases interrupt the translation of proteins from mRNA, which is why the entire phenomenon is called RNA interference.
What’s the takeaway? RNAi provides cells with a means of regulating gene expression after transcription.
4. Checking Understanding: Alternative Splicing and RNA Interference
The quiz below reviews what’s above, as well as some general features of post-transcriptional modification of RNA.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-M24_alternative splicing and RNA interference”] [h]
Labeled Diagrams: Alternative Splicing and RNA Interference
[i]
[q labels = “top”]
[l]introns
[f*] Excellent!
[fx] No. Please try again.
[l]mRNA
[f*] Correct!
[fx] No. Please try again.
[l]poly-A tail
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]pre-mRNA
[f*] Good!
[fx] No. Please try again.
[q labels = “top”]
[l]Alternative splicing
[f*] Great!
[fx] No. Please try again.
[l]pre-mRNA
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]transcription
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]translation
[f*] Great!
[fx] No. Please try again.
[q labels = “top”]
[l]miRNA
[f*] Excellent!
[fx] No. Please try again.
[l]mRNA degradation
[f*] Great!
[fx] No, that’s not correct. Please try again.
[l]DNA
[f*] Good!
[fx] No, that’s not correct. Please try again.
[l]precursor miRNA
[f*] Good!
[fx] No. Please try again.
[l]Blocked translation
[f*] Good!
[fx] No. Please try again.
[q multiple_choice=”true”] Which of the statements below most closely matches what’s being depicted in the following diagram?
[c]IEV2ZXJ5IEROQSBnZW5lIGhhcyBvbmUgY29ycmVzcG9uZGluZyBwcm90ZWluIHByb2R1Y3Qu[Qq]
[f]IE5vLiBMb29rIGF0ICYjODIyMDtELCYjODIyMTsgJiM4MjIwO0UsJiM4MjIxOyBhbmQgJiM4MjIwO0YsJiM4MjIxOyBhYm92ZS4gVGhyZWUgcHJvdGVpbnMgY29tZSBmcm9tIG9uZSBnZW5lLg==[Qq]
[c]IE11bHRpcGxlIHByb3RlaW5zIGNhbiBiZS Bwcm9kdWNlZCBmcm9tIG9uZSBnZW5lLg==[Qq]
[f]IEV4YWN0bHkuIEJ5IHNodWZmbGluZyBvciBkZWxldGluZyBleG9ucywgYWx0ZXJuYXRpdmUgc3BsaWNpbmcgYWxsb3dzIG11bHRpcGxlIHByb3RlaW5zIHRvIGJlIGNyZWF0ZWQgZnJvbSBvbmUgY29kaW5nIEROQS4=[Qq]
[c]IFByb2thcnlvdGVzIGhhdmUgbW9yZSBmbGV4aWJpbGl0eSBpbiBob3cgZ2VuZXMgYXJlIGV4cHJlc3NlZCB0aGFuIGRvIGV1a2FyeW90ZXM=[Qq]
[f]IE5vLiBUaGF0JiM4MjE3O3MgcHJvYmFibHkgbm90IHRydWUgKGp1c3QgY29uc2lkZXIgdGhlIGFsdGVybmF0aXZlIHNwbGljaW5nIHNob3duIGFib3ZlKSwgYW5kLCBpdCYjODIxNztzIG5vdCBkZXBpY3RlZCBpbiB0aGUgaW1hZ2UgYWJvdmUu[Qq]
[/qwiz]
5. Regulation of Translation by Proteins
In the previous sections, we saw how microRNAs can regulate translation. What follows are some additional processes by which translation can be regulated. We’ll look at this in the context of the regulation of cellular iron concentration.
Iron (Fe) is an essential nutrient. As a part of hemoglobin, it’s used to transport oxygen in red blood cells. Iron atoms also play a key role in the mitochondrial electron transport chain, as well as in the proteins involved in DNA replication and repair.
At the same time, too many free iron ions (in the form of Fe2+) can be toxic. To avoid iron toxicity, many organisms, including humans, use a protein called ferritin. Ferritin protein binds to iron, storing it in a non-toxic form (from which the iron can be released when the cell needs it).
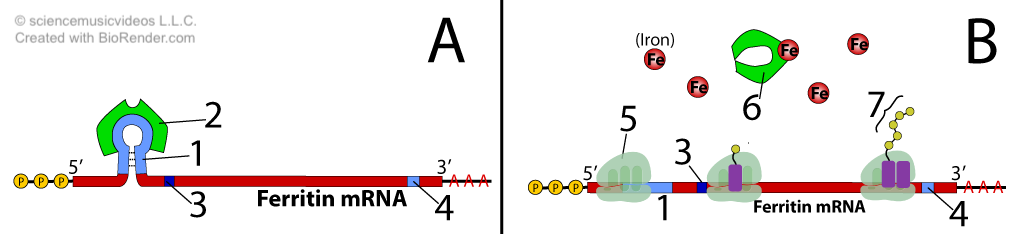
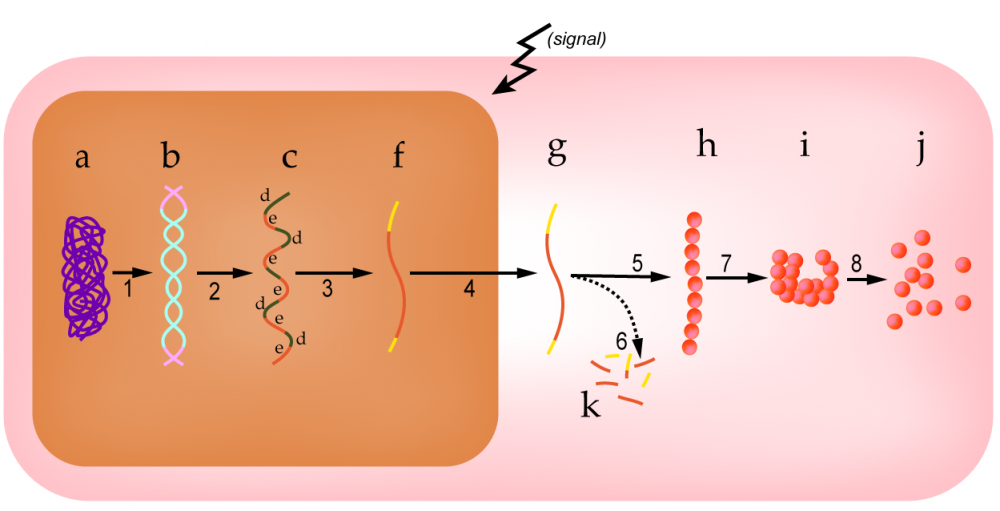
Ferritin mRNA contains a regulatory sequence that’s called an iron-response element (or “IRE,” shown at “1” in diagrams “A” and “B” above). When iron levels are low (diagram “A”), the IRE is bound by a regulatory protein called an IRE-binding protein, represented by “2.” When bound by this protein, the IRE blocks the start codon (“3”), preventing translation.
When iron levels are high (diagram “B”), iron binds with the IRE binding protein. This causes a conformational change that keeps the IRE-binding protein from binding with the iron response element (at “1”). Number “6” shows the IRE bound with iron. When iron binds with the IRE-binding protein, the iron response element sequence in the mRNA changes conformation. In diagram B, you can see that the mRNA has flattened out. As a result, ribosomes can bind with the mRNA and translate it, producing the ferritin protein (“7”). Note: number 4 in diagrams A and B above indicates the stop codon.
Note that this results in negative feedback: as ferritin is produced, more free iron will be captured, lowering the iron concentration.
The mechanism described above involves the control of translation by regulating the ability of ribosomes to bind with mRNA. Another mechanism of translational control involves mRNA degradation. The example of this that we’ll examine below also involves the regulation of iron metabolism, and it also involves an iron response element in mRNA. Note the overlap, and try to keep these two mechanisms separate as you study the example below.
The transferrin receptor protein is a membrane receptor that binds with transferrin, a glycoprotein that binds with iron in biological fluids. When transferrin binds with the transferrin receptor protein, it induces receptor-mediated endocytosis, creating a vesicle that brings the transferrin (and iron) into the cell.
Production of the transferrin receptor protein is also controlled by an IRE-binding protein.
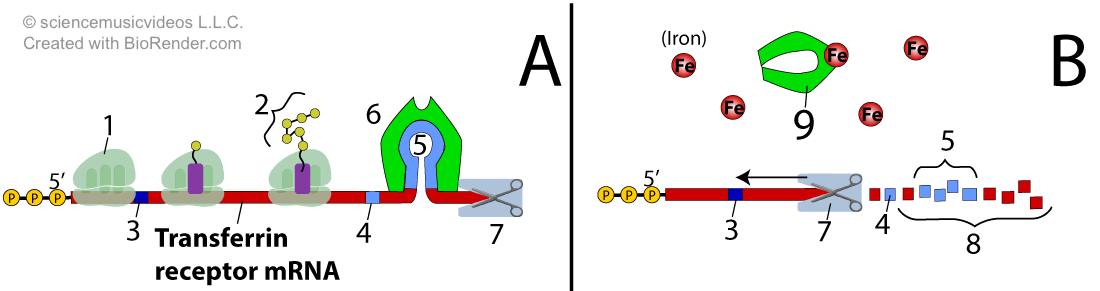
Diagram “A” shows what happens when iron levels are low. An IRE (iron response element) binding protein (shown at “6”) binds with the iron response element (“5”), a regulatory sequence in the untranslated region of the transferrin receptor protein mRNA. You can tell that the sequence is in the untranslated region because it’s on the 3′ end of the mRNA, after the stop codon (at “4”). The IRE/IRE-binding protein complex blocks degradative enzymes in the cytoplasm that would break down the mRNA. As a result, the mRNA is preserved, and ribosomes (1) can bind with the mRNA and translate the transferrin receptor protein (at “2”). This allows the cell to take in additional iron.
When iron levels are high, the cell no longer “wants” to take in additional iron (because, as stated above, high levels of iron can be toxic). Iron binds with the IRE binding protein, causing it to change shape in a way where it no longer binds to the IRE sequence (“5”) in the mRNA. This allows degradative enzymes to digest the mRNA (indicated by the broken-down RNA at “8”).
Again, note the negative feedback. High iron concentration keeps the mRNA for the transferrin receptor from being maintained in the cell. With the mRNA destroyed, little transferrin receptor will be produced, and little iron-containing transferrin will be brought into the cell, lowering the cellular iron level.
6. Protein Processing and Breakdown
A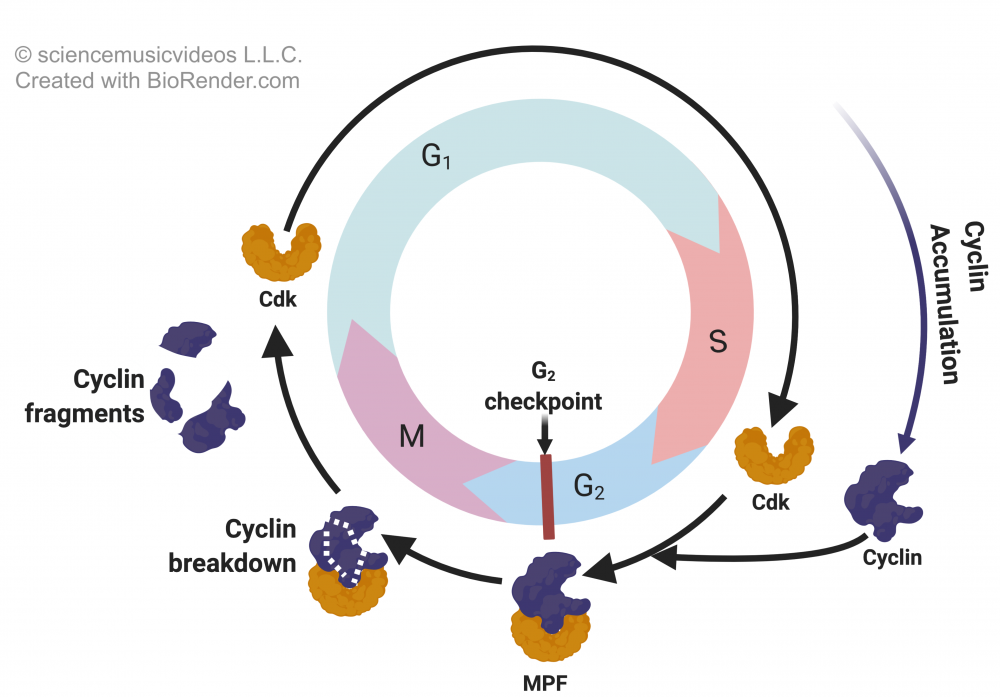 final opportunity for regulation of eukaryotic gene expression involves the modification and/or destruction of proteins. Think back for a moment to the cell cycle, which is controlled by the accumulation and then the destruction of cyclin. How is this destruction accomplished?
final opportunity for regulation of eukaryotic gene expression involves the modification and/or destruction of proteins. Think back for a moment to the cell cycle, which is controlled by the accumulation and then the destruction of cyclin. How is this destruction accomplished?
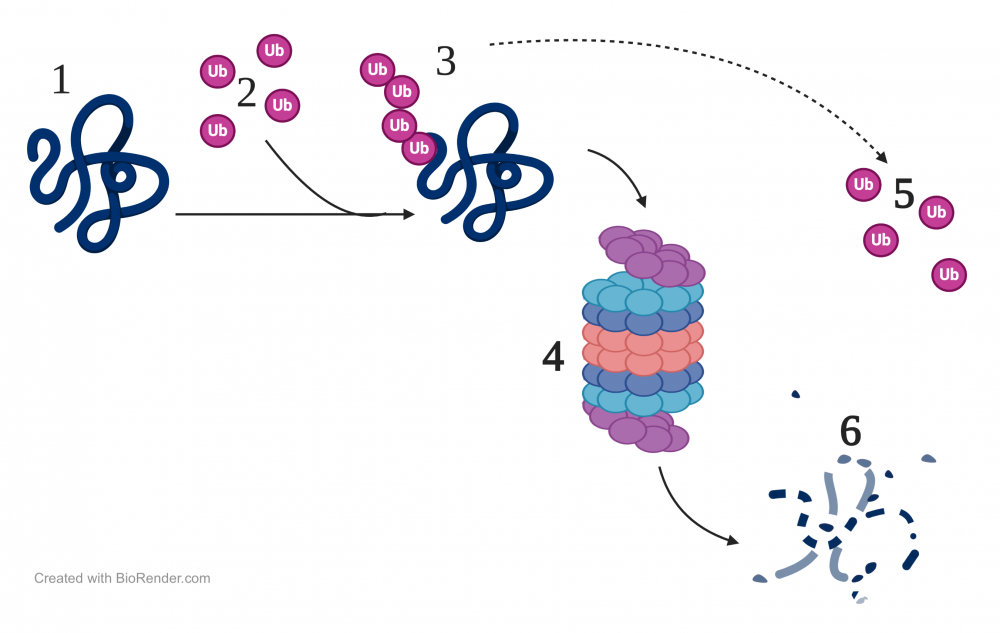
One method is to target a protein for destruction by tagging it with ubiquitin, a protein that is made up of 76 amino acids. Ubiquitin is present in cells throughout the eukaryotic domain (hence, it’s ubiquitous), and its structure is highly conserved in eukaryotic cells.
In the image to the right, a protein is shown at “1.” A team of ubiquitin-activating enzymes (not shown) attaches a short chain of ubiquitins to the protein (at “2”). Once tagged with ubiquitin (“3”), the protein is recognized by a proteasome (“4”). Proteasomes are protein complexes that break proteins down to short peptides (at “6”), which can then be recycled into individual amino acids that can be used in later instances of protein synthesis.
7. Eukaryotic Gene Regulation: Cumulative Quiz
That’s as far as we’re going to go in terms of explaining eukaryotic gene regulation. The following quiz will test your mastery of the material immediately above, and also include questions relevant to the entire module.
[qwiz random = “true” style=” width: 699px !important; ” qrecord_id=”sciencemusicvideosMeister1961-M24_Eukaryotic Gene Regulation, cumulative quiz”] [h]
Eukaryotic Gene regulation: Cumulative Quiz
[i]
[q] In the diagram below, the iron response element is at
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIGlyb24gcmVzcG9uc2UgZWxlbWVudCBpcyBhIHJlZ3VsYXRvcnkgc2VxdWVuY2UgaW4gdGhlIG1STkEsIGluZGljYXRlZCBieSAmIzgyMjA7MS4mIzgyMjE7[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgaXJvbiByZXNwb25zZSBlbGVtZW50IGlzIGEgcmVndWxhdG9yeSBzZXF1ZW5jZS4gTG9vayBmb3Igc29tZXRoaW5nIHRoYXQmIzgyMTc7cyBwYXJ0IG9mIHRoZSBETkEu[Qq]
[q] In the diagram below, the start codon is at
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IE5pY2UuIE51bWJlciAmIzgyMjA7MyYjODIyMTsgaXMgdGhlIHN0YXJ0IGNvZG9uLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBMb29rIGF0IGRpYWdyYW0gQi4gV2hlcmUmIzgyMTc7cyB0aGUgcG9pbnQgaW1tZWRpYXRlbHkgYWZ0ZXIgd2hpY2ggdGhlIHJpYm9zb21lIHN0YXJ0cyBwcm9kdWNpbmcgcHJvdGVpbnMu[Qq]
[q] In the diagram below, which is the system’s key regulatory protein, shown in its active form (where it prevents translation)?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gTnVtYmVyIDIgc2hvd3MgdGhlIGlyb24tcmVzcG9uc2UgZWxlbWVudCBiaW5kaW5nIHByb3RlaW4uIFdoZW4gYm91bmQgdG8gdGhlIElSRSwgdGhpcyBwcm90ZWluIHNodXRzIGRvd24gdHJhbnNsYXRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBXaGVuIGJvdW5kIHRvIHRoZSBJUkUsIHRoaXMgcHJvdGVpbiBzaHV0cyBkb3duIHRyYW5zbGF0aW9uLg==[Qq]
[q multiple_choice=”true”] The diagram below is about gene regulation through the control of
[c]IGFjZXR5bGF0aW9uIGFuZCBtZXRoeWxhdGlvbg==[Qq]
[f]IE5vLiBBY2V0eWxhdGlvbiBhbmQgbWV0aHlsYXRpb24gYXJlIHdheXMgdGhhdCBjZWxscyBjb250cm9sIEROQSBwYWNrYWdpbmcuIEF0IHRoaXMgcG9pbnQsIHdlIGhhdmUgbVJOQSBhbmQgcmlib3NvbWVzLiBDaG9vc2UgYSBsYXRlciBtb21lbnQgaW4gZ2VuZSByZWd1bGF0aW9uLg==[Qq]
[c]IGFsdGVybmF0aXZlIHNwbGljaW5n[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IE5vLiBBdCB0aGlzIHBvaW50LCB3ZSBoYXZlIG1STkEgYW5kIHJpYm9zb21lcy4gVHJhbnNjcmlwdGlvbiBoYXMgYWxyZWFkeSBoYXBwZW5lZC4=[Qq]
[c]IHRyYW5z bGF0aW9u[Qq]
[f]IEZhYnVsb3VzLiBBdCB0aGlzIHBvaW50LCB0aGVyZSBhcmUgbVJOQSBhbmQgcmlib3NvbWVzLiBUaGlzIGlzIGFib3V0IHRyYW5zbGF0aW9uYWwgY29udHJvbA==[Qq]
[q] In the diagram below, the regulatory sequence in the mRNA is at which number?
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IE5pY2Ugd29yayEgTnVtYmVyICYjODIyMDs1JiM4MjIxOyBpcyB0aGUgcmVndWxhdG9yIHNlcXVlbmNlIGluIHRoZSBtUk5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHBhcnQgb2YgdGhlIG1STkEgdGhhdCBzZWVtcyB0byBiZSBleGVydGluZyBzb21lIHJlZ3VsYXRvcnkgY29udHJvbCBvdmVyIG1STkEgZGVncmFkYXRpb24u[Qq]
[q] In the diagram below, regulation is about preventing or allowing enzymes that break down mRNA. These enzymes are indicated by which number?
[textentry single_char=”true”]
[c]ID c=[Qq]
[f]IEF3ZXNvbWUuIE51bWJlciAmIzgyMjA7NyYjODIyMTsgaW5kaWNhdGVzIGFuIGVuenltZSB0aGF0IGJyZWFrcyBkb3duIG1STkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBzb21ldGhpbmcgd2hpY2ggc2VlbXMgdG8gYmUgYnJlYWtpbmcgZG93biBtUk5BIGluICYjODIyMDtCLCYjODIyMTsgYnV0IGlzIGJsb2NrZWQgZnJvbSBkb2luZyBzbyBpbiBkaWFncmFtICYjODIyMDtBLiYjODIyMTs=[Qq]
[q] In the diagram below, which number shows a proteasome?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEV4Y2VsbGVudC4gTnVtYmVyIDQgc2hvd3MgYSBwcm90ZWFzb21lLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBQcm90ZWFzb21lcyBhcmUgbGFyZ2UgbW9sZWN1bGFyIGNvbXBsZXhlcyB0aGF0IGJyZWFrIGRvd24gcHJvdGVpbnMuIElmICYjODIyMDsxJiM4MjIxOyBpcyBhIHByb3RlaW4sIHRoZW4gd2hhdCBjb3VsZCBiZSBicmVha2luZyBpdCBkb3duPw==[Qq]
[q] In the diagram below, which number shows a protein that’s been tagged with ubiquitin for destruction?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IE5pY2Ugam9iISBOdW1iZXIgMyBzaG93cyBhIHByb3RlaW4gdGhhdCYjODIxNztzIGJlZW4gdGFnZ2VkIHdpdGggdWJpcXVpdGluLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBOdW1iZXIgJiM4MjIwOzEmIzgyMjE7IGlzIHRoZSBwcm90ZWluLCBhbmQgJiM4MjIwOzImIzgyMjE7IGlzIHViaXF1aXRpbi4=[Qq]
[q] In the diagram below, small peptides that can be recycled into amino acids are shown at
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]IEdvb2Qgd29yayEgJiM4MjIwOzYmIzgyMjE7IHNob3dzIHRoZSBwZXB0aWRlcyBsZWZ0IG92ZXIgYWZ0ZXIgdGhlIGxhcmdlciBwcm90ZWluIGhhcyBiZWVuIGJyb2tlbiBkb3duIGJ5IHRoZSBwcm90ZWFzb21lLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE0=[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBOdW1iZXIgJiM4MjIwOzEmIzgyMjE7IGlzIHRoZSBwcm90ZWluLCBhbmQgJiM4MjIwOzImIzgyMjE7IGlzIHViaXF1aXRpbi4=[Qq]
[q multiple_choice=”true”] Number 2 in the diagram below illustrates which concept that’s related to eukaryotic gene regulation?
[c]IGFjZXR5bGF0aW9uIGFuZCBtZXRoeWxhdGlvbg==[Qq]
[f]IE5vLiBBY2V0eWxhdGlvbiBhbmQgbWV0aHlsYXRpb24gYXJlIHdheXMgdGhhdCBjZWxscyBjb250cm9sIEROQSBwYWNrYWdpbmcuIFN0ZXAgMiBpcyBhYm91dCB3aGF0IGhhcHBlbnMgdG8gcHJlY3Vyc29yIFJOQSBhcyBpdCYjODIxNztzIG1hZGUgaW50byBtZXNzZW5nZXIgUk5BLg==[Qq]
[c]IGFsdGVybmF0aX ZlIHNwbGljaW5n[Qq]
[f]IEV4Y2VsbGVudCEgTnVtYmVyIDIgc2hvd3MgaG93IHRocm91Z2ggYWx0ZXJuYXRpdmUgc3BsaWNpbmcsIGEgc2luZ2xlIGdlbmUgY2FuIGJlIHRyYW5zZm9ybWVkIGludG8gdGhyZWUgcHJvdGVpbiBwcm9kdWN0cy4=[Qq]
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IE5vLiBXaGlsZSB0cmFuc2NyaXB0aW9uIGlzIHNob3duIGFib3ZlIChzdGVwIDEpLCB0aGUgcXVlc3Rpb24gaXMgYWJvdXQgc3RlcCAyLCB3aGVyZSBvbmUgcHJlLW1STkEgcmVzdWx0cyBpbiB0aHJlZSBkaWZmZXJlbnQgbVJOQXMsIGFuZCAzIGRpZmZlcmVudCBwcm90ZWlucy4gV2hhdCYjODIxNztzIHRoYXQgY2FsbGVkPw==[Qq]
[c]IHRyYW5zbGF0aW9uLg==[Qq]
[f]IE5vLiBUcmFuc2xhdGlvbiBpcyBoYXBwZW5pbmcgKGluIHN0ZXAgMyksIGJ1dCB0aGUgcXVlc3Rpb24gaXMgYWJvdXQgc3RlcCAyLCB3aGVyZSBvbmUgcHJlLW1STkEgcmVzdWx0cyBpbiB0aHJlZSBkaWZmZXJlbnQgbVJOQXMsIGFuZCAzIGRpZmZlcmVudCBwcm90ZWlucy4gV2hhdCYjODIxNztzIHRoYXQgcHJvY2VzcyBjYWxsZWQ/[Qq]
[q] In the diagram below, which letter indicates a precursor microRNA?
[textentry single_char=”true”]
[c]IE M=[Qq]
[f]IENvcnJlY3QhIExldHRlciAmIzgyMjA7YyYjODIyMTsgc2hvd3MgYSBwcmVjdXJzb3IgbWljcm9STkEuIFN1YnNlcXVlbnQgcHJvY2Vzc2luZyB3aWxsIHJlc3VsdCBpbiBhIG1pUk5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBzb21ldGhpbmcgdGhhdCYjODIxNztzIGEgcHJvZHVjdCBvZiB0cmFuc2NyaXB0aW9uLCBhbmQgd2hpY2ggd2lsbCBuZWVkIHN1YnNlcXVlbnQgcHJvY2Vzc2luZyB0byBiZWNvbWUgdGhlIG1pY3JvUk5BIHNob3duIGF0ICYjODIyMDtFLiYjODIyMTs=[Qq]
[q] In the diagram below, which letter shows a microRNA-associated protein?
[textentry single_char=”true”]
[c]IE k=[Qq]
[f]IE5pY2Ugam9iIS4gJiM4MjIwO0kmIzgyMjE7IHNob3dzIGEgbWljcm9STkEtYXNzb2NpYXRlZCBwcm90ZWluLCBhbHNvIGtub3duIGFzIGFu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHByb3RlaW4gdGhhdCBob29rcyBvbnRvIGEgc2luZ2xlLXN0cmFuZGVkIG1pY3JvUk5BLCBhbmQgd2hpY2ggKHdoZW4gd2l0aCB0aGUgbWlSTkEpIGNhbiBlaXRoZXIgYmxvY2sgdHJhbnNsYXRpb24gKEopIG9yIGRlZ3JhZGUgbVJOQSAoSyku[Qq]
[q] In one form of RNA interference, a complete match between a microRNA and a corresponding messenger RNA sequence sets the stage for subsequent enzymatic breakdown of the mRNA. In the diagram below, where do you see that breakdown happening?
[textentry single_char=”true”]
[c]IE s=[Qq]
[f]IEdvb2Qgd29yayEgTGV0dGVyICYjODIyMDtLJiM4MjIxOyBzaG93cyBtZXNzZW5nZXIgUk5BIGJlaW5nIGJyb2tlbiBkb3duIGFmdGVyIGJpbmRpbmcgd2l0aCBtaWNyb1JOQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBMb29rIGZvciBhIHBhcnQgb2YgdGhlIGltYWdlIHRoYXQgc2hvd3MgbVJOQSBiZWluZyBicm9rZW4gZG93biBpbnRvIGl0cyBjb21wb25lbnQgUk5BIG51Y2xlb3RpZGVzLg==[Qq]
[q multiple_choice=”true”] The diagram below shows a regulation of gene expression through
[c]IFJOQSBpbnRl cmZlcmVuY2U=[Qq]
[f]IEdyZWF0IGpvYi4gVGhlIGltYWdlIGFib3ZlIHNob3dzIFJOQSBpbnRlcmZlcmVuY2U=[Qq]
[c]IGFsdGVybmF0aXZlIHNwbGljaW5n[Qq]
[f]IE5vLiBXaGlsZSBpdCYjODIxNztzIHBvc3NpYmxlIHRoZSBhbHRlcm5hdGl2ZSBzcGxpY2luZyBjb3VsZCBoYXZlIHByb2R1Y2VkIHRoZSBtUk5BIGluIHRoaXMgZGlhZ3JhbSzCoCB0aGUga2V5IHBvaW50IGludm9sdmVzIG1vbGVjdWxlICYjODIyMDtFJiM4MjIxOyBpbnRlcmZlcmluZyB3aXRoIGEgcHJvY2VzcyB0aGF0IGludm9sdmVzIHJpYm9zb21lcyAoYXQgJiM4MjIwO0YmIzgyMjE7KS4gV2hhdCB0eXBlIG9mIG1vbGVjdWxlIGlzICYjODIyMDtFLCYjODIyMTsgYW5kIHdoYXQgcHJvY2VzcyBhcmUgcmlib3NvbWVzIGludm9sdmVkIHdpdGg/[Qq]
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[f]IE5vLiBXaGlsZSB0cmFuc2NyaXB0aW9uIGlzIHNob3duIGFib3ZlIChzdGVwIEIpLCB0aGUgcmVndWxhdG9yeSBwcm9jZXNzIGNvbWVzIGxhdGVyIGluIHRoZSBkaWFncmFtLiBIZXJlJiM4MjE3O3MgYSBoaW50OiB3aGF0IHR5cGUgb2YgbW9sZWN1bGUgaXMgJiM4MjIwO0UsJiM4MjIxOyBhbmQgd2hhdCBwcm9jZXNzIGFyZSByaWJvc29tZXMgKHNob3duIGF0ICYjODIyMDtGJiM4MjIxOykgaW52b2x2ZWQgd2l0aD8=[Qq]
[q] In the diagram below, which number is a DNA-bending protein?
[textentry single_char=”true”]
[c]ID E=[Qq]
[f]IE5pY2Ugam9iISAmIzgyMjA7MSYjODIyMTsgaXMgYSBETkEtYmVuZGluZyBwcm90ZWluLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBXaGljaCBwYXJ0IG9mIHRoZSBkaWFncmFtIHNlZW1zIHRvIGJlIGFzc29jaWF0ZWQgd2l0aCBhIGJlbmQgaW4gdGhlIEROQT8=[Qq]
[q] In the diagram below, which number is an enhancer?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IE5pY2Ugam9iISAmIzgyMjA7MiYjODIyMTsgaXMgYW4gZW5oYW5jZXIu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IFNvcnJ5LCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgZW5oYW5jZXIgaXMgcGFydCBvZiB0aGUgRE5BLCBhbmQgaXQmIzgyMTc7cyBjb21wb3NlZCBvZiAzIGNvbnRyb2wgZWxlbWVudHMu[Qq]
[q] In the diagram below, which number is pointing to one or more activators?
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IEF3ZXNvbWUhICYjODIyMDs0IGlzIHBvaW50aW5nIHRvIHR3byBhY3RpdmF0b3JzLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgYWN0aXZhdG9ycyBiaW5kIHdpdGggY29udHJvbCBlbGVtZW50cywgd2hpY2ggYXJlIHNlcXVlbmNlcyB3aXRoaW4gdGhlIEROQS4=[Qq]
[q] In the diagram below, the promoter is at
[textentry single_char=”true”]
[c]ID Y=[Qq]
[f]IFdheSB0byBnbyHCoCAmIzgyMjA7NiYjODIyMTsgcmVwcmVzZW50cyB0aGUgcHJvbW90ZXIu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgcHJvbW90ZXIgaXMganVzdCB1cHN0cmVhbSBvZiB0aGUgZ2VuZSwgYW5kIGl0JiM4MjE3O3Mgd2hlcmUgUk5BIHBvbHltZXJhc2UgYmluZHMu[Qq]
[q] In the diagram below, RNA polymerase is represented by
[textentry single_char=”true”]
[c]ID c=[Qq]
[f]IEV4Y2VsbGVudC7CoCAmIzgyMjA7NyYjODIyMTsgcmVwcmVzZW50cyBSTkEgcG9seW1lcmFzZS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBSTkEgcG9seW1lcmFzZSBiaW5kcyB3aXRoIHRoZSBwcm9tb3RlciwganVzdCB1cHN0cmVhbSBvZiB0aGUgZ2VuZS4=[Qq]
[q] In the diagram below, the TATA box is at
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIEEtVC1yaWNoIHJlZ2lvbiBhdCB0aGUgc3RhcnQgb2YgdGhlIHByb21vdGVyIGlzIGtub3duIGFzIHRoZSBUQVRBIGJveC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgVEFUQSBib3ggaXMgcmljaCBpbiBhZGVuaW5lIGFuZCB0aHltaW5lIGJhc2UgcGFpcnMu[Qq]
[q] In the diagram below, the template DNA is at
[textentry single_char=”true”]
[c]ID Q=[Qq]
[f]IE5pY2Ugam9iLiAmIzgyMjA7NCYjODIyMTsgaXMgdGhlIHRlbXBsYXRlIEROQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBUaGUgdGVtcGxhdGUgRE5BIGlzIHRoZSBzdHJhbmQgb2YgRE5BIHRoYXQmIzgyMTc7cyB0cmFuc2NyaWJlZCBpbnRvIFJOQS4gSWYgOCBpcyBSTkEsIHRoZW4gd2hhdCBoYXMgdG8gYmUgdGhlIHRlbXBsYXRlIHN0cmFuZCBvZiBETkE/[Qq]
[q multiple_choice=”true”] In the diagram below, which letter indicates an enhancer?
[c]IG E=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIGxldHRlciAmIzgyMjA7YSYjODIyMTsgaW5kaWNhdGVzIGFuIGVuaGFuY2VyLg==[Qq]
[c]IGM=[Qq]
[f]IE5vLCB0aGUgTGV0dGVyICYjODIyMDtjJiM4MjIxOyBpcyB0aGUgcHJvbW90ZXIuIEhlcmUmIzgyMTc7cyBhIGhpbnQuIEVuaGFuY2VycyBhcmUgcmVndWxhdG9yeSByZWdpb25zIHRoYXQgYXJlIHVzdWFsbHkgc29tZSBkaXN0YW5jZSBhd2F5IGZyb20gdGhlIGdlbmUu[Qq]
[c]IGQ=[Qq]
[f]IE5vLiBMZXR0ZXIgJiM4MjIwO2QmIzgyMjE7IGluZGljYXRlcyBhbiBleG9uLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBFbmhhbmNlcnMgYXJlIHJlZ3VsYXRvcnkgcmVnaW9ucyB0aGF0IGFyZSB1c3VhbGx5IHNvbWUgZGlzdGFuY2UgYXdheSBmcm9tIHRoZSBnZW5lLg==[Qq]
[c]ZQ==[Qq]
[f]Tm8uIExldHRlciAmIzgyMjA7ZSYjODIyMTsgaW5kaWNhdGVzIGFuIGludHJvbi4gSGVyZSYjODIxNztzIGEgaGludC4gRW5oYW5jZXJzIGFyZSByZWd1bGF0b3J5IHJlZ2lvbnMgdGhhdCBhcmUgdXN1YWxseSBzb21lIGRpc3RhbmNlIGF3YXkgZnJvbSB0aGUgZ2VuZS4=[Qq]
[q] In the diagram below, which letter indicates intron DNA?
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IE5pY2Ugam9iLiBUaGUgaW50cm9uIEROQSAoYXQgJiM4MjIwO2UmIzgyMjE7KSBpcyBETkEgdGhhdCBnZXRzIGN1dCBvdXQgb2YgdGhlIHByZS1STkEgYmVmb3JlIGl0IGdldHMgbWFkZSBpbnRvIG1STkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLsKgVGhlIGludHJvbiBETkEgaXMgRE5BIHRoYXQgZ2V0cyBjdXQgb3V0IG9mIHRoZSBwcmUtUk5BIGJlZm9yZSBpdCBnZXRzIG1hZGUgaW50byBtUk5BLiAmIzgyMjA7RiYjODIyMTsgcmVwcmVzZW50cyB0d28gUk5BIHNlZ21lbnRzIHRoYXQgaGF2ZSBiZWVuIGN1dCBvdXQmIzgyMzA7c28ganVzdCB3b3JrIHVwIGZyb20gdGhlcmUgYW5kIHlvdSYjODIxNztsbCBoYXZlIHRoZSBhbnN3ZXIuwqA=[Qq]
[q] In the diagram below, pre-mRNA (still with introns) is represented by which number?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEF3ZXNvbWUuICYjODIyMDsyJiM4MjIxOyBpcyBwcmUtbVJOQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTcGxpY2VkIG91dCBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBhdCAmIzgyMjA7Zi4mIzgyMjE7IA==TG9vayBmb3IgUk5BIHRoYXQgc3RpbGwgY29udGFpbnMgaW50cm9ucy7CoA==wqA=[Qq]
[q] In the diagram below, messenger RNA is represented by number
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEdvb2Qgd29yay4gJiM4MjIwOzMmIzgyMjE7IGlzIG1lc3NlbmdlciBSTkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBSTkEgaW50cm9ucyBjYW4gYmUgc2VlbiBpbiBudW1iZXLCoCAmIzgyMjA7Mi4mIzgyMjE7IA==TG9vayBmb3IgUk5BIGluIHdoaWNoIHRoZSBpbnRyb25zIGhhdmUgYmVlbiBzcGxpY2VkIG91dC4=[Qq]
[q multiple_choice=”true”] Which statement below best captures the significance of the process that’s represented in the diagram?
[c]IElmIHByb21vdGVycyBzaGFyZSBjb21tb24gY29udHJvbCBlbGVtZW50cywg dGhlbiBvbmUgc2lnbmFsIGNhbiBhY3RpdmF0ZSBtdWx0aXBsZSBnZW5lcy4=[Qq]
[f]IEV4Y2VsbGVudC4gV2hhdCYjODIxNztzIHNob3duIGFib3ZlIGlzIGhvdyBhbGwgdGhyZWUgZ2VuZXMgc2hhcmUgYSBjb21tb24gY29udHJvbCBlbGVtZW50IChpbmRpY2F0ZWQgYnkgJiM4MjIwO2MuJiM4MjIxOykgVGhpcyBhbGxvd3Mgb25lIHRyYW5zY3JpcHRpb24gZmFjdG9yICgmIzgyMjA7YiYjODIyMTspIHRvIHR1cm4gb24gYWxsIHRocmVlIGdlbmVzLg==[Qq]
[c]IFRocm91Z2ggYWx0ZXJuYXRpdmUgc3BsaWNpbmcsIG9uZSBnZW5lIGNhbiBwcm9kdWNlIG11bHRpcGxlIHByb3RlaW5zLg==[Qq]
[f]IE5vLiBUaGlzIGlzIGFib3V0IHRocmVlIGdlbmVzIHByb2R1Y2luZyB0aHJlZSBwcm90ZWlucyBhdCB0aGUgc2FtZSB0aW1lLiBXaGF0IGNvdWxkIGJyaW5nIHRoYXQgYWJvdXQ/[Qq]
[c]IFRoZSBjb21iaW5hdGlvbiBvZiBhY3RpdmF0b3JzIGluIGEgY2VsbCYjODIxNztzIG51Y2xldXMgZGV0ZXJtaW5lcyB3aGljaCBnZW5lcyB0aGF0IGNlbGwgd2lsbCBleHByZXNzLg==[Qq]
[f]IE5vLiBXaGlsZSB0aGF0JiM4MjE3O3MgdHJ1ZSwgaXQmIzgyMTc7cyBub3Qgd2hhdCYjODIxNztzIGJlaW5nIGlsbHVzdHJhdGVkIGFib3ZlLiBIZXJlIHlvdSBoYXZlIG9uZSBhY3RpdmF0b3IgYWN0aXZhdGluZyB0aHJlZSBnZW5lcy4gSG93IGNvdWxkIHRoYXQgY29tZSBhYm91dD8=[Qq]
[q multiple_choice=”true”] Which statement below best captures the significance of the process represented by the diagram?
[c]IEJpbmRpbmcgb2Ygc2lnbmFscyBhdCB0aGUgbWVtYnJhbmUgY2FuIHNldCBvZmYgYSBzaWduYWwgdHJhbnNkdWN0aW9uIGNhc2NhZGUsIHJlc3VsdGluZyBpbiBxdWljayBjaGFuZ2VzIHRocm91Z2hvdXQgdGhlIGNlbGwu[Qq]
[f]IE5vLiBUaGUgc2lnbmFsICgmIzgyMjA7ZiYjODIyMTspIGlzIGVudGVyaW5nIHRoZSBjZWxsJiM4MjE3O3MgY3l0b3BsYXNtLCBhbmQgbm90IGJpbmRpbmcgYXQgdGhlIG1lbWJyYW5lLiBOZXh0IHRpbWUsIGZpbmQgYSBiZXR0ZXIgZGVzY3JpcHRpb24gb2Ygd2hhdCYjODIxNztzIGhhcHBlbmluZy4=[Qq]
[c]IFRocm91Z2ggYWx0ZXJuYXRpdmUgc3BsaWNpbmcsIG9uZSBnZW5lIGNhbiBwcm9kdWNlIG11bHRpcGxlIHByb3RlaW5zLg==[Qq]
[f]IE5vLiBUaGlzIHNob3dzIG9uZSBnZW5lIGV4cHJlc3Npbmcgb25lIHByb3RlaW4gcHJvZHVjdC4gTmV4dCB0aW1lLCBmaW5kIGEgYmV0dGVyIGRlc2NyaXB0aW9uIG9mIHdoYXQmIzgyMTc7cyBoYXBwZW5pbmcu[Qq]
[c]IEhvcm1vbmVzIGNhbiBlbnRlciBjZWxscywgYmluZCB3aXRoIHJlY2 VwdG9ycywgYW5kIGJlY29tZSB0cmFuc2NyaXB0aW9uIGZhY3RvcnMu[Qq]
[f]IEZhYnVsb3VzLiBUaGlzIGlsbHVzdHJhdGlvbiBpcyBzaG93aW5nIGhvdyBzdGVyb2lkIGhvcm1vbmVzIGxpa2UgZXN0cm9nZW4gd29yay4gVGhleSBlbnRlciBjZWxscywgYmluZCB3aXRoIHJlY2VwdG9ycywgYW5kIHRoZW4gYWN0IGFzIHRyYW5zY3JpcHRpb24gZmFjdG9ycy4=[Qq]
[q] In the diagram below, a proteasome would be active at number
[textentry single_char=”true”]
[c]ID g=[Qq]
[f]IE5pY2UuIFByb3RlYXNvbWVzIGJyZWFrIGRvd24gcHJvdGVpbnMu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBQcm90ZWFzb21lcyBicmVhayBkb3duIHByb3RlaW5zLg==[Qq]
[q] In the diagram below, alternative RNA splicing would occur at which number?
[textentry single_char=”true”]
[c]ID M=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwOzMmIzgyMjE7IHNob3dzIHdoZXJlIGEgcHJlLVJOQSB0cmFuc2NyaXB0IGlzIHByb2Nlc3NlZCB0byBjcmVhdGUgbVJOQS4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBUaGVyZSBhcmUgdHdvIHJlcHJlc2VudGF0aW9ucyBvZiBSTkEgaW4gdGhlIGNlbGwmIzgyMTc7cyBudWNsZXVzLiBMb29rIHRoZXJlLCBhbmQgb25lIG9mIHRoZSBhcnJvd3Mgd2lsbCBiZSB5b3VyIGFuc3dlci4=[Qq]
[q] In the diagram below, heterochromatin is indicated by which letter?
[textentry single_char=”true”]
[c]IG E=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIGxldHRlciAmIzgyMjA7YSYjODIyMTsgc2hvd3MgZGVuc2VseSBwYWNrZWQgY2hyb21hdGluLCBhbHNvIGtub3duIGFzIGhldGVyb2Nocm9tYXRpbi4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50OiBGaW5kIHdoZXJlIHRoZSBETkEgaXMgdGlnaHRseSBwYWNrZWQgYW5kIHRoZXJlZm9yZSB1bmF2YWlsYWJsZSBmb3IgdHJhbnNjcmlwdGlvbi4=[Qq]
[q] In the diagram below, “1” is [hangman].
[c]IGhldGVyb2Nocm9tYXRpbg==[Qq]
[f]IEdvb2Qh[Qq]
[q] In the diagram below, “3” is a structure composed of eight [hangman] proteins. The entire structure is called a [hangman]
.
[c]IGhpc3RvbmU=[Qq]
[f]IEdvb2Qh[Qq]
[c]IG51Y2xlb3NvbWU=[Qq]
[f]IEdyZWF0IQ==[Qq]
[q] The key difference between the DNA at “1” and “2” is that the DNA at “1” is inaccessible to RNA[hangman], and therefore can’t be [hangman]
.
[c]IHBvbHltZXJhc2U=[Qq]
[f]IENvcnJlY3Qh[Qq]
[c]IHRyYW5zY3JpYmVk[Qq]
[f]IEV4Y2VsbGVudCE=[Qq]
[q] The illustration at left is designed to depict female________________. Microscopic examination of the cells of this person’s (or any female’s) nuclei would reveal that one of her X chromosomes had been transformed into a ________________ body, a tight piece of ________________ that is not expressed. Because X chromosomes are inactivated in cells at ________________, there are patches of cells throughout the body that express the alleles in one of the X chromosomes, but not the other.
[l] Barr
[fx] No. Please try again.
[f*] Correct!
[l] heterochromatin
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[l] mosaicism
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[l] random
[fx] No, that’s not correct. Please try again.
[f*] Correct!
[q] In the diagram below, “E” (along with “G” and “H” shows [hangman]. The heavy blue line underneath the ribosome (at “F”) is [hangman]. One way to describe what’s being shown below is RNA [hangman]
.
[c]IG1pUk5B[Qq]
[c]IG1STkE=[Qq]
[c]IGludGVyZmVyZW5jZQ==[Qq]
[q] In the diagram below, “8” is pointing to a(n) [hangman] group. As a result of “8,” the gene indicated by “7” is available for[hangman].
[c]IGFjZXR5bA==[Qq]
[c]IHRyYW5zY3JpcHRpb24=[Qq]
[q]In the diagram below, “b” is pointing to a segment of RNA that isn’t in the mRNA (at “a”). Therefore, “b” must be an [hangman].
[c]aW50cm9u[Qq]
[q multiple_choice=”true”] The gene controlling the development of the pelvic spine is the Pitx1 gene, which codes for the Pitx1 protein. When the genes of fish with and without pelvic spines were compared, it was found that
[c]IFRoZSBzZXF1 ZW5jZSBvZiA=UGl0eDE=IHdhcyB0aGUgc2FtZSBpbiBmaXNoIHdpdGggcGVsdmljIHNwaW5lcyBhbmQgZmlzaCB3aXRob3V0IHBlbHZpYyBzcGluZXM=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIChzdXJwcmlzaW5nKSBmaW5kaW5nIHdhcyB0aGF0IHRoZXJlIHdhcyBubyBkaWZmZXJlbmNlIGluIHRoZSBjb2Rpbmcgc2VxdWVuY2UgYmV0d2VlbiB0aGUgdHdvIHR5cGVzIG9mIGZpc2gu[Qq]
[c]IFRoZSBzZXF1ZW5jZSBvZiA=UGl0eDE=IHdhcyBub3JtYWwgaW4gZmlzaCB3aXRoIHBlbHZpYyBzcGluZXMsIGJ1dCBmaXNoIHdpdGggbm8gcGVsdmljIHNwaW5lcyBoYWQgYSByZWFkaW5nIGZyYW1lIG11dGF0aW9uIHRoYXQgbWFkZSB0aGVpciA=UGl0eDE=IGdlbmUgbm9uLWZ1bmN0aW9uYWwu[Qq]
[f]IE5vLiBUaGUgUGl0eDE=IHNlcXVlbmNlIHdhcyB0aGUgc2FtZSBpbiBmaXNoIHdpdGggYSBwZWx2aWMgc3BpbmUgYW5kIHRob3NlIHdpdGhvdXQgYSBwZWx2aWMgc3BpbmUu[Qq]
[c]IFRoZSBzZXF1ZW5jZSBvZiA=UGl0eDE=IHdhcyBub3JtYWwgaW4gZmlzaCB3aXRob3V0IHNwaW5lcywgYnV0IGZpc2ggd2l0aCBzcGluZXMgaGFkIGEgbXV0YXRlZCB2ZXJzaW9uIG9mIHRoZSBnZW5lLsKg[Qq]
[f]IE5vLiBUaGUgUGl0eDE=IHNlcXVlbmNlIHdhcyB0aGUgc2FtZSBpbiBmaXNoIHdpdGggYSBwZWx2aWMgc3BpbmUgYW5kIHRob3NlIHdpdGhvdXQgYSBwZWx2aWMgc3BpbmUu[Qq]
[q labels=”top”]Label the diagram below.
[l]enhancers/switches
[fx] No. Please try again.
[f*] Excellent!
[l]mRNA
[fx] No. Please try again.
[f*] Correct!
[l]protein-coding region
[fx] No. Please try again.
[f*] Correct!
[l]promoter
[fx] No. Please try again.
[f*] Excellent!
[l]RNA polymerase
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[x][restart]
[/qwiz]
Links
This tutorial ends this module on eukaryotic gene expression.
- Return to the Eukaryotic Gene Expression Menu
- Use the menu choices above to choose another topic.
- Proceed to Animal Development and Cell Specialization (the next module in many AP/College curricula).