1. Introduction
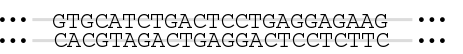
Sequencing is taking a sample of DNA – anything from a small fragment to the entire genome of an organism – and figuring out the specific sequence of A, T, C, and G nucleotides that make it up. This tutorial will explain one method by which that can be done.
The explanation that follows will be a lot easier if you’ve already done my previous tutorial about the polymerase chain reaction (because the two processes have a lot in common).
2. Sanger Sequencing Concept 1: Dideoxynucleotides
The first widely used sequencing method was developed by Frederick Sanger in 1977. This method, called Sanger Sequencing, earned Sanger the 1980 Nobel Prize and was the basis of the techniques used to sequence the entire human genome, a feat that was completed in 2001 as the culmination of the Human Genome Project.
Sanger’s method involves in vitro DNA synthesis using dideoxynucleotides. In vitro means “in a test tube,” as opposed to in a living cell. The prefix “dideoxy” means “deoxygenated twice.” That’s because the monomers of DNA, deoxyribonucleotides (or just “deoxynucleotides”) have been deoxygenated once: they have one less oxygen atom than ribonucleotides, the monomer of RNA.
To see this, take a look at molecule “B” below and find the number 2. That “2” represents a carbon atom in deoxyribose (the sugar in DNA), and notice that it has a hydrogen atom attached to it. In ribonucleotides (see molecule “C”), you’d find an –OH in that spot. So, deoxyribose is deoxygenated ribose. Now, look at molecule “A,” which is a dideoxynucleotide. Notice that the number 3 carbon atom has a hydrogen atom attached to it. Compare it to the number 3 carbon in deoxyribose, which has an –OH attached to it. Hence, molecule “A” has been deoxygenated twice (compared to ribose). It’s a dideoxynucleotide.
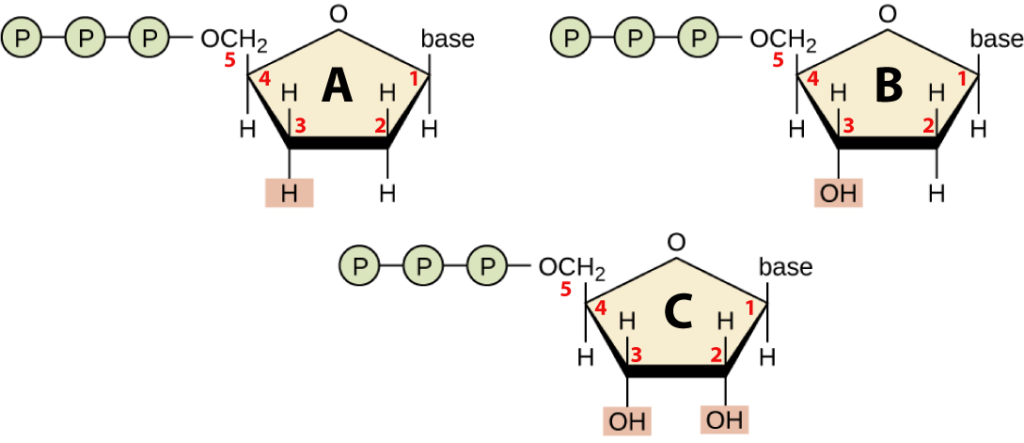
Finally, note that all of the molecules above have three phosphate groups attached to them. When the nucleic acid polymers DNA and RNA are synthesized, two of these phosphate groups will be broken off and will form bonds with water molecules. That reaction liberates the free energy that can do important work for the cell: creating DNA or RNA.
3. Sanger Sequencing Concept 2: incorporating ddNucleotides during DNA replication causes chain termination
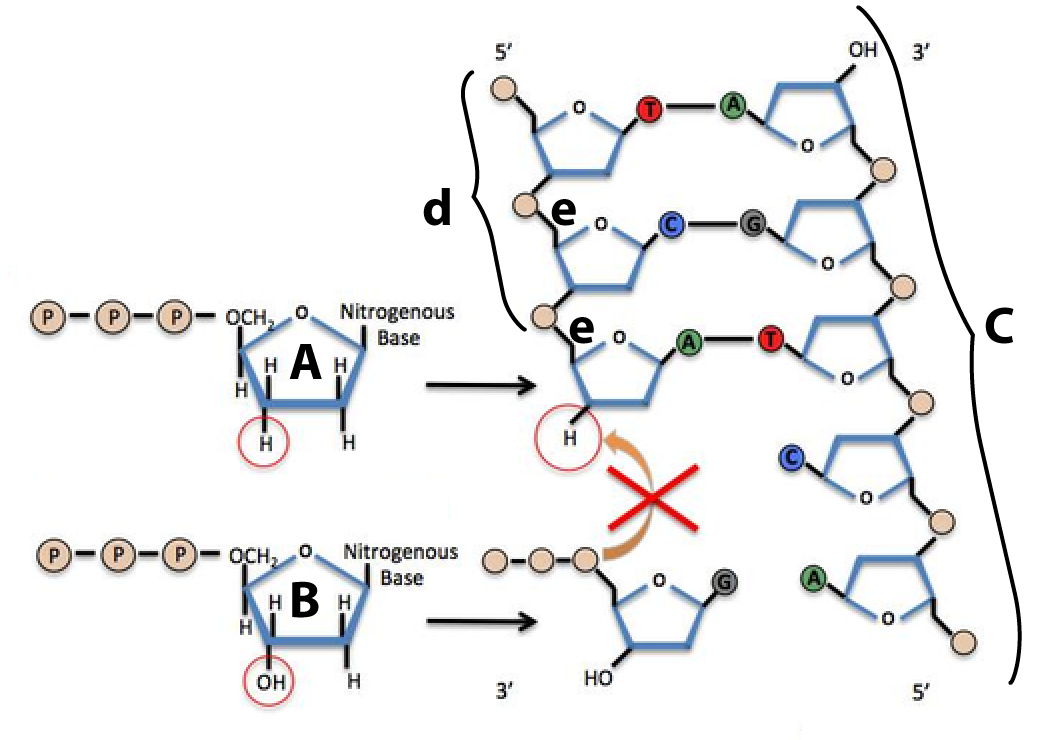
The chemical difference between deoxynucleotides and dideoxynucleotides is at the heart of Sanger sequencing. That’s because when dideoxynucleotides (molecule “A,” at right) are incorporated into a newly synthesized DNA strand (d) by DNA polymerase (which is not shown), they can’t form a sugar-phosphate bond (e) between the dideoxynucleotide and the next nucleotide (b) at the 3′ end of the growing strand. That causes chain termination. As we’ll see in the next section, that chain termination is the basis of the Sanger sequencing method. But first, let’s check to make sure you’ve gotten everything so far.
4. Sanger Sequencing Quiz 1
[qwiz random = “true” qrecord_id=”sciencemusicvideosMeister1961-Sanger Sequencing 1 (ddNucleotides), M17″] [h]
Sanger sequencing quiz 1
[i]
[q] Figuring out the order of the nucleotide bases in DNA is called[hangman].
[c]IHNlcXVlbmNpbmc=[Qq]
[f]IENvcnJlY3Qh[Qq]
[q] In the diagram below, the dideoxynucleotide is shown at letter
[textentry single_char=”true”]
[c]IE E=[Qq]
[f]IEdvb2QgSm9iISAmIzgyMjA7QSYjODIyMTsgaXMgYSBkaWRlb3h5bnVjbGVvdGlkZS7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiDCoFdoZW4gY29tcGFyZWQgdG8gcmlib3NlIChhdCAmIzgyMjA7QyYjODIyMTspLCA=ZGlkZW94eQ==bnVjbGVvdGlkZXMgaGF2ZSBsb3N0IHR3byBveHlnZW4gYXRvbXMuIFdoaWNoIG9mIHRoZSBtb2xlY3VsZXMgc2hvd24gY291bGQgYmUgYSA=ZGlkZW94eQ==bnVjbGVvdGlkZT8=[Qq]
[q] In the diagram below, the type of nucleotide that you’d find in messenger RNA is
[textentry single_char=”true”]
[c]IE M=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0MmIzgyMjE7IHJlcHJlc2VudHMgYSByaWJvbnVjbGVvdGlkZSwgdGhlIG1vbm9tZXIgb2YgUk5BLg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgbW9ub21lcnMgb2YgUk5BIGFyZSByaWJvbnVjbGVvdGlkZXMuIFRoZSBzdWdhciB0aGF0IG1ha2VzIHRoZW0gdXAgaXMgcmlib3NlLCBhbmQgaXQgaGFzIHR3byBoeWRyb3h5bHMgKOKAkyBPSCBncm91cHMpLsKg[Qq]
[q] In the diagram below, the type of nucleotide that you’d find in the DNA making up your cells is
[textentry single_char=”true”]
[c]IE I=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0ImIzgyMjE7IHJlcHJlc2VudHMgYSBkZW94eXJpYm9udWNsZW90aWRlLCB0aGUgbW9ub21lciBvZiBETkEu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgbW9ub21lcnMgb2YgRE5BIGFyZSBkZW94eXJpYm9udWNsZW90aWRlcy4gVGhleSYjODIxNzt2ZSBsb3N0IG9uZSBveHlnZW4gYXRvbSBjb21wYXJlZCB0byByaWJvc2UuIFlvdSBjYW4gZmluZCByaWJvc2UgYnkgZmluZGluZyB0aGUgbnVjbGVvdGlkZSB0aGF0IGhhcyB0d28gaHlkcm94eWwgZ3JvdXBzLsKg[Qq]
[q] In the diagram below, the type of nucleotide used by Sanger to bring about chain termination is
[textentry single_char=”true”]
[c]IE E=[Qq]
[f]IE5pY2Ugam9iLiAmIzgyMjA7QSYjODIyMTsgcmVwcmVzZW50cyBhIGRpZGVveHludWNsZW90aWRlLCBhbmQgdGhlc2UgYnJpbmcgYWJvdXQgY2hhaW4gdGVybWluYXRpb24u[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBEaWRlb3h5bnVjbGVvdGlkZXMgZG9uJiM4MjE3O3QgaGF2ZSBoeWRyb3h5bCBncm91cHMgYXR0YWNoZWQgdG8gdGhlaXIgMyYjODI0MjsgY2FyYm9uLCBhbmQgdGhlcmVmb3JlIGNhbiYjODIxNzt0IGZvcm0gc3VnYXItcGhvc3BoYXRlIGJvbmRzLsKg[Qq]
[q] In the diagram below, the template strand is shown at
[textentry single_char=”true”]
[c]IE M=[Qq]
[f]IEV4Y2VsbGVudC4gJiM4MjIwO0MmIzgyMjE7IHJlcHJlc2VudHMgdGhlIHRlbXBsYXRlIHN0cmFuZC7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgdGVtcGxhdGUgaXMgdGhlIHN0cmFuZCB0aGF0JiM4MjE3O3MgJiM4MjIwO3JlYWQmIzgyMjE7IGJ5IEROQSBwb2x5bWVyYXNlIGFzIGl0IHN5bnRoZXNpemVzIGEgbmV3IHN0cmFuZCBpbiB0aGUgNSYjODI0MjsgdG8gMyYjODI0MjsgZGlyZWN0aW9uLsKg[Qq]
[q] In the diagram below, a dideoxynucleotide is shown at
[textentry single_char=”true”]
[c]IE E=[Qq]
[f]IE5pY2Ugam9iLiAmIzgyMjA7QSYjODIyMTsgcmVwcmVzZW50cyBhIGRpZGVveHludWNsZW90aWRlLsKgwqA=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBIZXJlJiM4MjE3O3MgYSBoaW50LiBUaGUgaW5jb3Jwb3JhdGlvbiBvZiBhIGRpZGVveHludWNsZW90aWRlIGludG8gYSBncm93aW5nIEROQSBzdHJhbmQgcmVzdWx0cyBpbiBzdHJhbmQgdGVybWluYXRpb24uwqA=[Qq]
[q] In the diagram below, sugar-phosphate bonds are shown
[textentry single_char=”true”]
[c]IG U=[Qq]
[f]IFdheSB0byBnbyEgTGV0dGVyICYjODIyMDtlJiM4MjIxOyByZXByZXNlbnRzIHN1Z2FyLXBob3NwaGF0ZSBib25kcy7CoA==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBTdWdhci1waG9zcGhhdGUgYm9uZHMgYXJlIHRoZSBib25kcyB0aGF0IGNvbm5lY3QgYWRqYWNlbnQgbnVjbGVvdGlkZXMuwqA=[Qq]
[q] Incorporation of a dideoxynucleotide into a growing DNA strand (as shown below results in chain [hangman].
[c]IHRlcm1pbmF0aW9u[Qq]
[q] Dideoxynucleotides can’t form sugar-[hangman] bonds at their 3′ end, resulting in chain termination.
[c]IHBob3NwaGF0ZQ==[Qq]
[/qwiz]
5. Sanger Sequencing Concept 3: How ddnucleotide chain termination reveals DNA sequences
Pretend that you’re Frederick Sanger, trying to figure out the sequence of a strand of DNA that’s 9 nucleotides long. To assist your learning, I’m going to tell you what the sequence is, but the point is that by following the steps below, you could figure out the sequence (which is what Sanger did).
The sequence you’re interested in is shown in blue.

Sanger wasn’t concerned about 5′ ACAA 3′, shown in black. That’s because those nucleotides represent known primer binding sites, which I’ll explain more about below.
What Sanger did was clone many copies of this sequence, using gene cloning techniques that you can learn about in the first tutorial in this module about genetic engineering. After 1983 this type of cloning could have been done using PCR.
With lots of DNA in hand, Sanger then divided the DNA into four fractions, which he placed into four separate reaction tubes. Each tube contained the DNA that Sanger was trying to sequence. Each tube also contained many copies of each of the necessary ingredients. These were:
- A primer. This is a short stretch of DNA that will bind to the primer binding site. In this case, the primer 3′ TGTT 5′ will bind with 5′ ACAA 3′. The primer was created with radioactive nucleotides so that the DNA fragments it would bind with could be visualized by exposing them to an X-ray film after Southern blotting.
- DNA polymerase. This is the key enzyme in DNA replication
- Free deoxynucleotides. A generalized formula for one of these is shown in Figure 2 above, at “B.” Sanger would have added deoxynucleotides with each of the four bases to each tube: dATP (with adenine), dTTP (with thymine), dGTP (with guanine) and dCTP with cytosine). Note two things: 1) “dATP,” “dTTP” and so on indicate the normal deoxyribonucleotide that gets used during DNA synthesis. 2) the letters “TP” indicate that each nucleotide has three phosphate groups attached to it, enabling it to be incorporated by DNA polymerase into a new strand. For example, molecule “B” in figure 3 above is guanine triphosphate or dGTP.
Each tube would contain a different type of dideoxynucleotide. This is the form of nucleotide that is doubly deoxygenated, which causes chain termination. Each tube contained one of the following: ddATP (with adenine), ddTTP (with thymine), ddGTP (with guanine), and ddCTP with cytosine).
Here’s what the whole setup would have looked like.

Now, let’s try to imagine what would have happened in tube # 4, the one with ddCTP.
The primer would bind with the primer binding site at the start of the template strand, at the 3′ end of the template.
When DNA Polymerase got to the first G (immediately after the primer) it could insert ddCTP. That would result in the shortest possible fragment: just one nucleotide past the primer, as shown below.
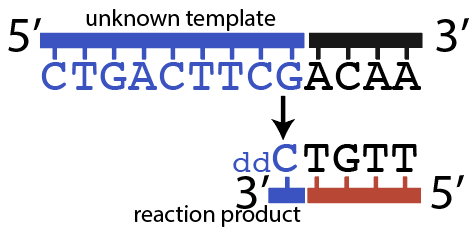
But it’s also possible that DNA polymerase would have inserted a dCTP (the “normal” nucleotide), and continued elongating the new strand until it got to the 7th nucleotide in the template strand, which is the 2nd “G.” At that point, DNA polymerase might insert another ddCTP, which would look like this:
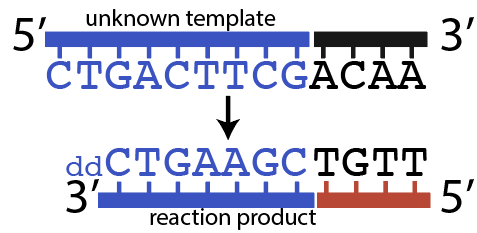
6. Chain Termination, Checking Understanding
If you’re getting it, you should be able to predict what will happen in tube number 3, the tube with ddGTP. Answer the questions below.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Sanger Sequencing 2 (chain termination)M17″] [h]
Chain Termination in Reaction Tube 3 (with ddGTP): Checking Understanding
[q multiple_choice=”true”] You’re doing Sanger sequencing on the template 5’CTGACTTCG3′. In the reaction tube that has ddGTP, how many fragments will be produced?
[c]IDHigIPigIM=[Qq][c]IDLigIPigIM=[Qq][c]IDPigI PigIM=[Qq][c]IDQ=
Cg==[Qq][f]IE5vLiBUaGUgdW5rbm93biB0ZW1wbGF0ZSBpc8KgIDXigJlDVEdBQ1RUQ0cz4oCyLiBJbiB0aGUgZGRHVFAgdHViZSwgdGhlIGRkR1RQIG1vbGVjdWxlcyB3aWxsIGJlIGluY29ycG9yYXRlZCBpbnRvIHRoZSBuZXdseSBzeW50aGVzaXplZCBzdHJhbmQgd2hlbmV2ZXIgdGhlcmUmIzgyMTc7cyBhICYjODIyMDtDJiM4MjIxOyAoY3l0b3NpbmUpIGluIHRoZSB0ZW1wbGF0ZS4gSG93IG1hbnkgQ3MgYXJlIHRoZXJlPw==[Qq]
[f]IE5vLiBUaGUgdW5rbm93biB0ZW1wbGF0ZSBpc8KgIDXigJlDVEdBQ1RUQ0cz4oCyLiBJbiB0aGUgZGRHVFAgdHViZSwgdGhlIGRkR1RQIG1vbGVjdWxlcyB3aWxsIGJlIGluY29ycG9yYXRlZCBpbnRvIHRoZSBuZXdseSBzeW50aGVzaXplZCBzdHJhbmQgd2hlbmV2ZXIgdGhlcmUmIzgyMTc7cyBhICYjODIyMDtDJiM4MjIxOyAoY3l0b3NpbmUpIGluIHRoZSB0ZW1wbGF0ZS4gSG93IG1hbnkgQ3MgYXJlIHRoZXJlIGluIHRoZSB0ZW1wbGF0ZT8=[Qq]
[f]IEV4Y2VsbGVudC4gVGhlIHVua25vd24gdGVtcGxhdGUgaXPCoCA14oCZQ1RHQUNUVENHM+KAsi4gSW4gdGhlIGRkR1RQIHR1YmUsIHRoZSBkZEdUUCBtb2xlY3VsZXMgd2lsbCBiZSBpbmNvcnBvcmF0ZWQgaW50byB0aGUgbmV3bHkgc3ludGhlc2l6ZWQgc3RyYW5kIHdoZW5ldmVyIHRoZXJlJiM4MjE3O3MgYSAmIzgyMjA7QyYjODIyMTsgKGN5dG9zaW5lKSBpbiB0aGUgdGVtcGxhdGUuIFRoZXJlIGFyZSAzIGN5dG9zaW5lcywgc28gdGhlcmUgd2lsbCBiZSAzIGZyYWdtZW50cyBwcm9kdWNlZC4=[Qq]
[f]IE5vLiBUaGUgdW5rbm93biB0ZW1wbGF0ZSBpc8KgIDXigJlDVEdBQ1RUQ0cz4oCyLiBJbiB0aGUgZGRHVFAgdHViZSwgdGhlIGRkR1RQIG1vbGVjdWxlcyB3aWxsIGJlIGluY29ycG9yYXRlZCBpbnRvIHRoZSBuZXdseSBzeW50aGVzaXplZCBzdHJhbmQgd2hlbmV2ZXIgdGhlcmUmIzgyMTc7cyBhICYjODIyMDtDJiM4MjIxOyAoY3l0b3NpbmUpIGluIHRoZSB0ZW1wbGF0ZS4gSG93IG1hbnkgQ3MgYXJlIHRoZXJlPw==[Qq]
[q] You’re doing Sanger sequencing on the template 5’CTGACTTCG3′. In the reaction tube that has ddGTP, the smallest fragment will be how many nucleotides long?
[textentry single_char=”true”]
[c]ID I=[Qq]
[f]IEV4Y2VsbGVudCEgVGhlIHNtYWxsZXN0IGZyYWdtZW50IHdpbGwgYmUgdHdvIG51Y2xlb3RpZGVzIGxvbmcuIFRoYXQmIzgyMTc7cyBiZWNhdXNlIHRoZSBmaXJzdCBjeXRvc2luZSBpcyB0d28gbnVjbGVvdGlkZXMgYXdheSBmcm9tIHRoZSAzJiM4MjQyOyBlbmQgb2YgdGhlIHRlbXBsYXRlIHN0cmFuZCwgYW5kIHRoYXQmIzgyMTc7cyB3aGVyZSBETkEgcG9seW1lcmFzZSB3aWxsIGluc2VydCB0aGUgZmlyc3QgZGRHVFA=Lg==
Cg==[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBnb2luZyB0byByZWFkIHRoZSB0ZW1wbGF0ZSBETkEgaW4gdGhlIDMmIzgyNDI7IHRvIDUmIzgyNDI7IGRpcmVjdGlvbiBhbmQgbGF5IGRvd24gbmV3IGNvbXBsZW1lbnRhcnkgbnVjbGVvdGlkZXMgYXMgaXQgc3ludGhlc2l6ZXMgYSBuZXcgc3RyYW5kLiBJbiByZWFjdGlvbiB0dWJlIDMsIHdpdGggZGRHIG51Y2xlb3RpZGVzLCBETkEgcG9seW1lcmFzZSBpcyBnb2luZyB0byBhcnJpdmUgYXQgdGhlIGZpcnN0IEMgKGN5dG9zaW5lKSB0aGF0IGl0IGVuY291bnRlcnMsIGFuZCBwdXQgaW4gYSBjb21wbGVtZW50YXJ5IGRHVFAgKGd1YW5pbmUpLiBIb3dldmVyLCBiZWNhdXNlIHRoZSByZWFjdGlvbiB0dWJlIGhhcyBkZEdUUCBudWNsZW90aWRlcywgRE5BIHBvbHltZXJhc2UgbWlnaHQgaW5zZXJ0IGEgZGRHVFAsIHJlc3VsdGluZyBpbiBjaGFpbiB0ZXJtaW5hdGlvbi4gSWYgeW91IGNvdW50IGhvdyBmYXIgdGhhdCBmaXJzdCBDIGlzIGZyb20gdGhlIDMmIzgyNDI7IGVuZCBvZiB0aGUgdGVtcGxhdGUsIHlvdSYjODIxNztsbCBrbm93IGhvdyBsb25nIHRoYXQgc21hbGxlc3QgZGRHVFAtdGVybWluYXRlZCBmcmFnbWVudCB3aWxsIGJlLg==[Qq]
[q] You’re doing Sanger sequencing on the template 5’CTGACTTCG3′. In the reaction tube that has ddGTP, the medium-sized fragment will be how many nucleotides long?
[textentry single_char=”true”]
[c]ID U=[Qq]
[f]IE5pY2UhIFRoZSBtZWRpdW0gZnJhZ21lbnQgd2lsbCBiZSBmaXZlIG51Y2xlb3RpZGVzIGxvbmcuIFRoYXQmIzgyMTc7cyBiZWNhdXNlIHRoZSBzZWNvbmQgY3l0b3NpbmUgaXMgZml2ZSBudWNsZW90aWRlcyBhd2F5IGZyb20gdGhlIDMmIzgyNDI7IGVuZCBvZiB0aGUgdGVtcGxhdGUgc3RyYW5kLCBhbmQgdGhhdCYjODIxNztzIHdoZXJlIEROQSBwb2x5bWVyYXNlIHdpbGwgaW5zZXJ0IHRoZSBzZWNvbmQgZGRHVFAu[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLCB0aGF0JiM4MjE3O3Mgbm90IGNvcnJlY3Qu[Qq]
[c]ICo=[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBnb2luZyB0byByZWFkIHRoZSB0ZW1wbGF0ZSBETkEgaW4gdGhlIDMmIzgyNDI7IHRvIDUmIzgyNDI7IGRpcmVjdGlvbiBhbmQgbGF5IGRvd24gbmV3IGNvbXBsZW1lbnRhcnkgbnVjbGVvdGlkZXMgYXMgaXQgYnVpbGRzIGEgbmV3IHN0cmFuZC4gSW4gcmVhY3Rpb24gdHViZSAzLCB3aXRoIGRkR1RQIG51Y2xlb3RpZGVzLCBETkEgcG9seW1lcmFzZSBpcyBnb2luZyB0byBhcnJpdmUgYXQgdGhlIGZpcnN0IEMgKGN5dG9zaW5lKSB0aGF0IGl0IGVuY291bnRlcnMsIGFuZCBwdXQgaW4gYSBjb21wbGVtZW50YXJ5IEcgKGd1YW5pbmUpLiBBc3N1bWUsIGluIHRoaXMgY2FzZSwgdGhhdCBETkEgcG9seW1lcmFzZSBpbnNlcnRzIGEgbm9ybWFsIEcgbnVjbGVvdGlkZSAoYSBkR1RQKSB3aGVyZSB0aGUgZmlyc3QgY3l0b3NpbmUgaXMgb24gdGhlIHRlbXBsYXRlIEROQSwgYW5kIHRoZW4gbW92ZXMgb24gdG8gdGhlIHNlY29uZCBvbmUuIEF0IHRoaXMgc2Vjb25kIHBvc2l0aW9uLCBETkEgcG9seW1lcmFzZSBtaWdodCBpbnNlcnQgYSBkZEdUUCwgcmVzdWx0aW5nIGluIGNoYWluIHRlcm1pbmF0aW9uLiBJZiB5b3UgY291bnQgaG93IGZhciB0aGF0IHNlY29uZCBDIGlzIGZyb20gdGhlIDMmIzgyNDI7IGVuZCBvZiB0aGUgdGVtcGxhdGUsIHlvdSYjODIxNztsbCBrbm93IGhvdyBsb25nIHRoYXQgbWVkaXVtLXNpemVkIGRkR1RQLXRlcm1pbmF0ZWQgZnJhZ21lbnQgd2lsbCBiZS4=
Cg==Tm90ZSB0aGF0IHRoZXJlIHdpbGwgYmUgdGhyZWUgZGlmZmVyZW50IGZyYWdtZW50cyBvZiB0aHJlZSBkaWZmZXJlbnQgbGVuZ3RocywgZGVwZW5kaW5nIG9uIHdoZXJlIGEgZGRHVFAgaXMgZmlyc3QgaW5zZXJ0ZWQu[Qq]
[q] You’re doing Sanger sequencing on the template 5’CTGACTTCG3′. In the reaction tube that has ddGTP, the largest fragment will be how many nucleotides long?
[textentry single_char=”true”]
[c]ID k=[Qq]
[f]IE5pY2UhIFRoZSBsYXJnZXN0IGZyYWdtZW50IHdpbGwgYmUgbmluZSBudWNsZW90aWRlcyBsb25nLiBUaGF0JiM4MjE3O3MgYmVjYXVzZSB0aGUgdGhpcmQgY3l0b3NpbmUgaXMgbmluZSBudWNsZW90aWRlcyBhd2F5IGZyb20gdGhlIDMmIzgyNDI7IGVuZCBvZiB0aGUgdGVtcGxhdGUgc3RyYW5kLCBhbmQgdGhhdCYjODIxNztzIHdoZXJlIEROQSBwb2x5bWVyYXNlIHdpbGwgaW5zZXJ0IHRoZSB0aGlyZCBkZEdUUC4=[Qq]
[c]IEVudGVyIHdvcmQ=[Qq]
[f]IE5vLg==[Qq]
[c]ICo=[Qq]
[f]IE5vLiBETkEgcG9seW1lcmFzZSBpcyBnb2luZyB0byByZWFkIHRoZSB0ZW1wbGF0ZSBETkEgaW4gdGhlIDMmIzgyNDI7IHRvIDUmIzgyNDI7IGRpcmVjdGlvbiBhbmQgbGF5IGRvd24gbmV3IGNvbXBsZW1lbnRhcnkgbnVjbGVvdGlkZXMuIEFzc3VtZSwgaW4gdGhpcyBjYXNlLCB0aGF0IEROQSBwb2x5bWVyYXNlIGluc2VydHMgYSBub3JtYWwgRyBudWNsZW90aWRlIHdoZXJlIHRoZSBmaXJzdCBhbmQgc2Vjb25kIGN5dG9zaW5lcyBhcmUgb24gdGhlIHRlbXBsYXRlIEROQSwgYW5kIHRoZW4gbW92ZXMgb24gdG8gdGhlIHRoaXJkIG9uZS4gQXQgdGhpcyB0aGlyZCBwb3NpdGlvbiwgRE5BIHBvbHltZXJhc2UgaW5zZXJ0cyBhIGRkR1RQLCByZXN1bHRpbmcgaW4gY2hhaW4gdGVybWluYXRpb24uIElmIHlvdSBjb3VudCBob3cgZmFyIHRoYXQgdGhpcmQgQyBpcyBmcm9tIHRoZSAzJiM4MjQyOyBlbmQgb2YgdGhlIHRlbXBsYXRlLCB5b3UmIzgyMTc7bGwga25vdyBob3cgbG9uZyB0aGF0IGxhcmdlc3QgZGRHVFAtdGVybWluYXRlZCBmcmFnbWVudCB3aWxsIGJlLg==[Qq]
[/qwiz]
SUMMARY
[qwiz]
[h]Reaction tube 3 (ddGTP)
[q]In tube number 3, the tube with ddGTP, you’d get three possible reaction products. Click “show the answer” to reveal what they are:
[c]U2hvdyB0aGUgYW5zd2Vy[Qq]
[f]
Cg==[Qq]
[/qwiz]
Now, look one more time at the unknown template, and try to imagine the reaction products in tube 1 (with ddATP) and tube 2 (with ddTTP). You might want to sketch out your ideas on a sheet of paper before moving on to the next step.

7. Sanger Sequencing Concept 3: Electrophoresis of Reaction Products and Analysis of Results
Sanger’s next move was to take the reaction products in each tube and separate them by size by gel electrophoresis. If you need a refresher of how that works, consult my tutorial about DNA fingerprinting by clicking here. Otherwise, just remember that the smallest fragments will move the most.
Remember that Sanger didn’t know the sequence: he was figuring it out. What he did know was that by using dideoxynucleotides, he would get nucleotide chains that terminated with the ddnucleotide in that test tube.
What you’re trying to do is to predict the way the DNA segments produced in each reaction chamber would sort themselves out by size. Based on our results above, I’ll do the first one (the results from the ddCTP tube) for you. There were two segments produced: one was one nucleotide long (not including the primer) and one was seven nucleotides long.
 |
 |
Both of these fragments will start in the well near the negative pole of the gel. The fragment with one nucleotide will migrate to fragment length row 1. That’s because that fragment is (not including the primer) only one nucleotide long. The fragment that’s seven nucleotides long will migrate to fragment length row 7. You do the rest (and don’t be afraid to guess).
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Sanger Sequencing 3, Electrophoresis (M17)”]
[h]Sanger Sequencing: Analysis of Results of Electrophoresis
[q labels = “top”]The unknown sequence is 5’CTGACTTCG3′. Drag the fragments that end with ddnucleotides into the right place on the virtual gel below. Note that just for this exercise, the modified nucleotides are abbreviated as “ddA,” “ddT,” “ddG,” and “ddC.”
| Negative pole | ||||
| ddATP well | ddTTP well | ddGTP well | ddCTP well | Fragment Length |
| _________ | _________ | _________ | _________ | 9 |
| _________ | _________ | _________ | _________ | 8 |
| _________ | _________ | _________ | ____ddC__ | 7 |
| _________ | _________ | _________ | _________ | 6 |
| _________ | _________ | _________ | _________ | 5 |
| _________ | _________ | _________ | _________ | 4 |
| _________ | _________ | _________ | _________ | 3 |
| _________ | _________ | _________ | _________ | 2 |
| _______ | _________ | _________ | ___ddC__ | 1 |
[l]ddA
[fx] No, that’s not correct. Please try again.
[f*] Great!
[l]ddT
[fx] No. Please try again.
[f*] Correct!
[l]ddG
[fx] No, that’s not correct. Please try again.
[f*] Excellent!
[q labels = “bottom”]What Sanger saw after electrophoresis and Southern Blotting was something like the gel shown below. You know that DNA is always synthesized in the 5′ to 3′ direction. Therefore, you know that in the newly synthesized strands, the longest fragments are going to be the ones with a ddnucleotide at the 3′ end, and the shortest will have a ddnucleotide at the 5′ end. So, now your job is to use the gel to figure out the sequence of the newly synthesized strand. Once you’ve done that, just use the base pairing rules to determine the sequence of the original strand. I’ve given you two hints (so try not to look above on this page).
[l]A
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]T
[fx] No, that’s not correct. Please try again.
[f*] Good!
[l]C
[fx] No. Please try again.
[f*] Great!
[l]G
[fx] No. Please try again.
[f*] Great!
[/qwiz]
8. Sanger Sequencing: Application
That’s it for understanding Sanger Sequencing. To recap:
- Sanger used dideoxynucleotides to bring about chain termination during in vitro DNA synthesis.
- Electrophoresis of the products in each reaction tube would sort the terminated chains by size.
- Analysis of the position of the fragments on the gel would reveal the sequence. The nucleotide at the 3′ end of the longest newly synthesized fragment would be complementary to the first nucleotide on the 5′ end of the unknown template. The next longest fragment would end with the complement of the next nucleotide, and so on.
Try predicting and analyzing a few Sanger sequencing gels.
[qwiz qrecord_id=”sciencemusicvideosMeister1961-Sanger Sequencing 3, Practice (M17)”] [h]
Sanger Sequencing Practice
[q labels = “bottom”]Here’s a sequence of DNA: 5’TTAGACCCGAT 3′. Drag the bar below (representing a DNA fragment) into the right place on the virtual gel below. Note the order of the reaction tubes (A, T, C, G). Just to be clear: the “___” is the DNA fragment that you’re dragging into the correct position.
| Negative pole | ||||
| ddATP well | ddTTP well | ddCTP well | ddGTP well | Fragment Length |
| _________ | _________ | _________ | _________ | 11 |
| _________ | _________ | _________ | _________ | 10 |
| _________ | _________ | _________ | _________ | 9 |
| _________ | _________ | _________ | _________ | 8 |
| _________ | _________ | _________ | _________ | 7 |
| _________ | _________ | _________ | _________ | 6 |
| _________ | _________ | _________ | _________ | 5 |
| _________ | _________ | _________ | _________ | 4 |
| _________ | _________ | _________ | _________ | 3 |
| _________ | _________ | _________ | _________ | 2 |
| ________ | _________ | _________ | _________ | 1 |
| Positive Pole | ||||
[l]____
[f*] Good!
[fx] No. Please try again.
[q labels = “bottom”]Sanger sequencing of an unknown strand of DNA produces the gel shown below. What is the sequence of the newly synthesized strand, and what is the sequence of the original DNA template?
Newly synthesized DNA:
3′_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ 5′
Template DNA (what you’re sequencing)
5′_ _ _ _ _ _ _ _ _ _ _ _ _ _ _ _ 3′
[l]A
[f*] Great!
[fx] No. Please try again.
[l]T
[f*] Correct!
[fx] No. Please try again.
[l]C
[f*] Excellent!
[fx] No, that’s not correct. Please try again.
[l]G
[f*] Great!
[fx] No. Please try again.
[q multiple_choice=”true”] An unknown fragment of DNA is being processed for Sanger sequencing. Electrophoresis results in the gel below. What’s the sequence of this fragment of DNA?
[c]IDUmIzgyNDI7IEFUR0NDQUdUQSAzJiM4MjQyOw==[Qq]
[f]SGVyZSYjODIxNztzIGEgaGludDogVGhlIGxvbmdlc3QgZnJhZ21lbnQgKHdoaWNoIGlzIGF0IHRoZSB0b3ApIGVuZHMgaW4gQS4gVGhhdCBtZWFucyBpdCB3YXMgc3ludGhlc2l6ZWQgZnJvbSBhIHBpZWNlIG9mIEROQSB3aGVyZSB0aGUgZmlyc3QgbnVjbGVvdGlkZSB3YXMgNSAmIzgyMTY7VC4=[Qq]
[c]IDUmIzgyNDI7IFRBQ0dHVENBVCAzJiM4MjE3Ow==[Qq]
[f]SGVyZSYjODIxNztzIGEgaGludDogVGhlIGxvbmdlc3QgZnJhZ21lbnQgKHdoaWNoIGlzIGF0IHRoZSB0b3ApIGVuZHMgaW4gQS4gVGhhdCBtZWFucyBpdCB3YXMgc3ludGhlc2l6ZWQgZnJvbSBhIHBpZWNlIG9mIEROQSB3aGVyZSB0aGUgZmlyc3QgbnVjbGVvdGlkZSB3YXMgNSAmIzgyMTY7VC4gVGhlIHNlY29uZCBsb25nZXN0IGZyYWdtZW50IGVuZGVkIHdpdGggVC4gVGhhdCBtZWFucyBpdCB3YXMgc3ludGhlc2l6ZWQgZnJvbSBhIHBpZWNlIG9mIEROQSB3aGVyZSB0aGUgc2Vjb25kIG51Y2xlb3RpZGUgd2FzIEEuIEtlZXAgb24gdXNpbmcgdGhlIHNhbWUgbWV0aG9kLg==[Qq]
[c]IDUmIzgyNDI7IFRBQ1RH R0NBVCAzJiM4MjQyOw==[Qq]
[f]RXhjZWxsZW50LiBUaGUgZ2VsIGFib3ZlIHdvdWxkIGJlIGNyZWF0ZWQgZHVyaW5nIHRoZSBzZXF1ZW5jaW5nIG9mIDUmIzgyMTc7VEFDVEdHQ0FUIDMmIzgyMTc7Lg==[Qq]
[q] An unknown fragment of DNA is being processed for Sanger sequencing. Electrophoresis results in the gel below. What’s the sequence of this fragment of DNA?
[c]NSYjODI0MjsgQUNDR1RBR0FUIDMmIzgyNDI7[Qq]
[f]Tm8uIFJlYWRpbmcgZnJvbSB0aGUgdG9wIGRvd24sIHlvdSBjYW4gc2VlIHRoYXQgdGhlIGxvbmdlc3Qgc3ludGhlc2l6ZWQgc3RyYW5kIGVuZGVkIGluIEEuIFRoYXQgbWVhbnMgdGhhdCBUIGlzIHRoZSBmaXJzdCBudWNsZW90aWRlIG9uIHRoZSA1JiM4MjQyOyBzaWRlLsKgIFlvdSB0YWtlIGl0IGZyb20gdGhlcmUu[Qq]
[c]NSYjODI0MjsgVEdHQ0 FUQ1RBIDMmIzgyNDI7[Qq]
[f]RXhjZWxsZW50Lg==[Qq]
[c]IDUmIzgyNDI7IFRBR0FUR0NDQSYjODIxNzs=[Qq]
[f]Tm8uIFJlYWRpbmcgZnJvbSB0aGUgdG9wIGRvd24sIHlvdSBjYW4gc2VlIHRoYXQgdGhlIGxvbmdlc3Qgc3ludGhlc2l6ZWQgc3RyYW5kIGVuZGVkIGluIEEuIFRoYXQgbWVhbnMgdGhhdCBUIGlzIHRoZSBmaXJzdCBudWNsZW90aWRlIG9uIHRoZSA1JiM4MjQyOyBzaWRlLiBUaGUgbmV4dCBsb25nZXN0IHN5bnRoZXNpemVkIHN0cmFuZCBlbmRlZCBpbiBDLiBUaGF0IG1lYW5zIHRoYXQgRyB3YXMgdGhlIG5leHQgbnVjbGVvdGlkZS4gWW91IHRha2UgaXQgZnJvbSB0aGVyZS4=[Qq]
[/qwiz]
9. Sequencing Advances: Sanger Method and Beyond
Sanger Sequencing is still in use today, though in an automated form that’s replaced four-lane electrophoresis with capillary gel electrophoresis.
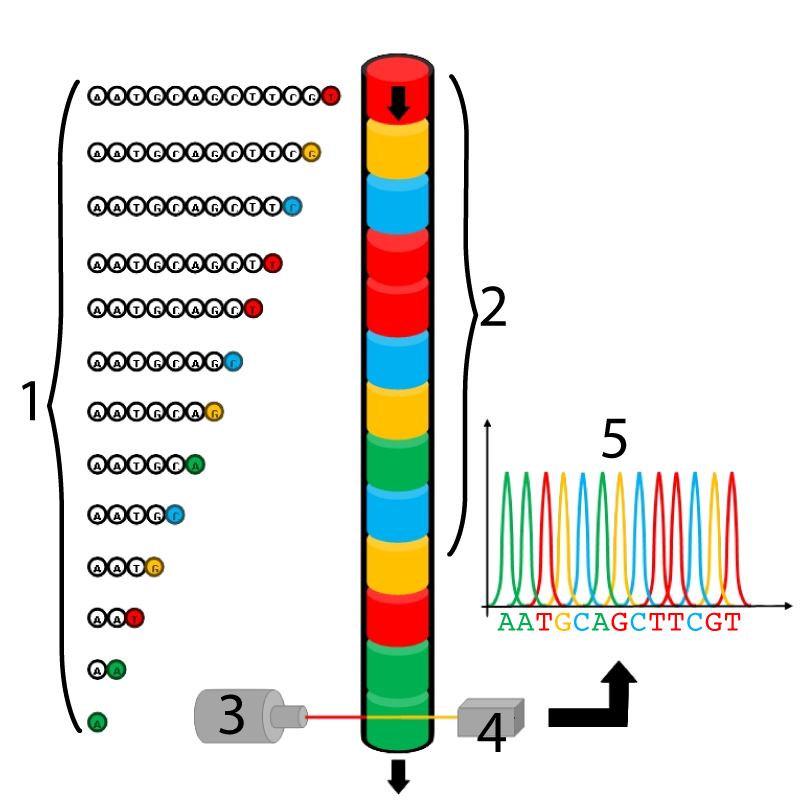
The process begins in much the same way as with Sanger’s original method: using ddnucleotides and in vitro DNA synthesis, fragments are created, each of which represents the position of a specific nucleotide. The ddnucleotides used in capillary gel electrophoresis, however, each have a special fluorescent pigment molecule attached, enabling them to be identified later. After fragment synthesis (step 1), all the fragments are fed into a gel-containing capillary tube (2). Electrophoresis happens within that tube, sorting the fragments by size. The fragments flow past a laser beam (3), which stimulates each ddnucleotide to fluoresce with a unique color. This color is read by a detector (4) and the sequence is recorded on a computer.
Those advances, however, are just the start of advances in sequencing. Sanger sequencing, while still in use, is being supplanted by high-throughput techniques. If you’re interested, you can read about these on this page on Wikipedia, or you can even take this course in Next Generation sequencing offered by the European Bioinformatics Institute.
Links
At this point in your study of biology, the information on sequencing above is all you need to know. Click the links below to continue
- Gene Editing Using CRISPR-Cas9 (the next tutorial in this series)
- Genetic Engineering and Biotechnology Main Menu

